Pharmacophore-Conditioned Diffusion Model for Ligand-Based De Novo Drug Design
Pharmacophore-Conditioned Diffusion Model for Ligand-Based De Novo Drug Design
Amira Alakhdar, Barnabas Poczos, Newell Washburn
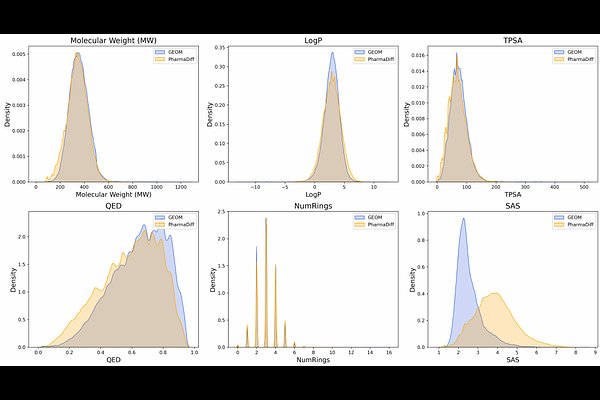
AbstractDeveloping bioactive molecules remains a central, time- and cost-heavy challenge in drug discovery, particularly for novel targets lacking structural or functional data. Pharmacophore modeling presents an alternative for capturing the key features required for molecular bioactivity against a biological target. In this work, we present PharmaDiff, a pharmacophore-conditioned diffusion model for 3D molecular generation. PharmaDiff employs a transformer-based architecture to integrate an atom-based representation of the 3D pharmacophore into the generative process, enabling the precise generation of 3D molecular graphs that align with predefined pharmacophore hypotheses. Through comprehensive testing, PharmaDiff demonstrates superior performance in matching 3D pharmacophore constraints compared to ligand-based drug design methods. Additionally, it achieves higher docking scores across a range of proteins in structure-based drug design, without the need for target protein structures. By integrating pharmacophore modeling with 3D generative techniques, PharmaDiff offers a powerful and flexible framework for rational drug design.