Unearthing the rhizosphere microbiome recruited by ancestral bread wheat landraces
Unearthing the rhizosphere microbiome recruited by ancestral bread wheat landraces
HERNANDEZ SORIANO, M. C.; Warren, F. J.; Hildebrand, F.; Wingen, L. U.; Miller, A. J.; Griffiths, S.
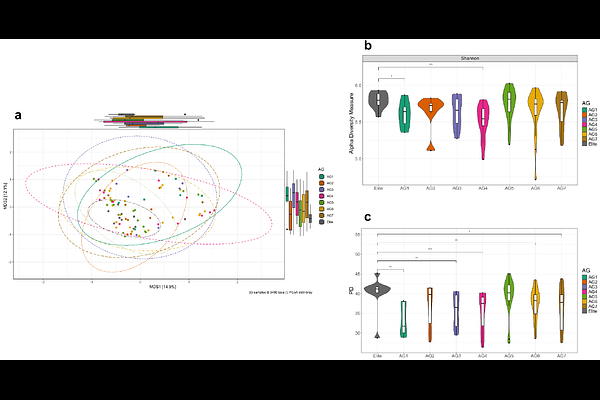
AbstractCrop root traits that modulate the soil microbiome can turn the tide of reduced fertility in intensively farmed land by optimising nutrient availability and resilience to environmental stresses. Advantageous genetic diversity that allows adaptation to nutrient availability is present in historic crop genotypes. The Watkins collection of bread wheat landraces is an unexploited resource, carrying untapped phenotypic traits. Here, we show that the rhizosphere microbiome assembly of these landraces is distinct compared to elite varieties, specifically those that come from ancestral groups (AGs) not used in modern breeding. We used 16S rRNA sequencing to identify changes in microbial communities of rhizosphere soil collected from 81 landraces and two elite varieties. We found high similarity in microbiome recruitment between the elite cultivars and the two AGs genetically closest to the elite. The rhizosphere microbiome of five AGs genetically distant from the elite cultivars showed significant differences in the abundance of taxa involved in nitrogen and carbon turnover, keystone taxa and associations within the microbial network. Our findings suggest that genes to recruit or suppress microbial taxa in the rhizosphere are shared by landraces from these AGs. Selective breeding for traits to control microbial functions can enhance soil productivity and crop performance.