Integration of diploid T2T parental genomes with progeny genomes and transcriptomes identifies resistance genes in the grape powdery mildew resistance loci Ren6 and Ren7 of Vitis piazeskii
Integration of diploid T2T parental genomes with progeny genomes and transcriptomes identifies resistance genes in the grape powdery mildew resistance loci Ren6 and Ren7 of Vitis piazeskii
Massonnet, M.; Figueroa-Balderas, R.; Cochetel, N.; Riaz, S.; Pap, D.; Walker, M. A.; Cantu, D.
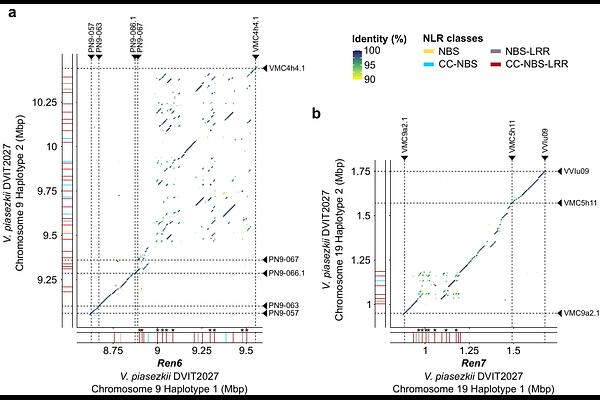
AbstractThe Chinese grape accession Vitis piasezkii DVIT2027 carries two loci associated with powdery mildew (PM) resistance, Ren6 and Ren7. Although these loci differ in the timing and strength of their response to Erysiphe necator, both are consistent with recognition by intracellular immune receptors. Here, we aimed to identify the nucleotide-binding leucine-rich repeat (NLR) genes responsible for PM resistance within Ren6 and Ren7. To do so, we assembled diploid telomere-to-telomere (T2T) genomes of V. piasezkii DVIT2027 and V. vinifera F2-35, the resistant and susceptible parents, respectively, of a segregating F1 population. We then integrated these assemblies with deep resequencing data from eight F1 sib-lines carrying different combinations of Ren6 and Ren7, and generated trio-binned, parent-phased genomes for six of them. This allowed us to resolve and phase the Ren6 and Ren7 loci and their PM-susceptible alternative haplotypes. Comparative analyses revealed extensive structural variation, particularly duplications, and numerous short polymorphisms between resistant and susceptible haplotypes, leading to complete sequence specificity among all NLRs in Ren6 and Ren7, with several lacking allelic counterparts in their respective susceptible haplotypes. Expression profiling across PM-resistant sib-lines identified four and two candidate CC-NBS-LRR genes associated with Ren6 and Ren7, respectively, based on both expression and haplotype specificity. These results provide a high-resolution view of two key PM resistance loci and support the identification of candidate resistance genes for functional validation and targeted grapevine breeding.