BTRR complex deficiency is a driver for genomic instability in Bloom syndrome
BTRR complex deficiency is a driver for genomic instability in Bloom syndrome
Gönenc, I. I.; Wolff, A.; Busley, A. V.; Wieland, A.; Tijhuis, A.; Müller, C.; Wardenaar, R.; Argyriou, L.; Kaulfuss, S.; Räschle, M.; Spierings, D. C. J.; Foijer, F.; Bastians, H.; Yigit, G.; Zibat, A.; Cyganek, L.; Wollnik, B.
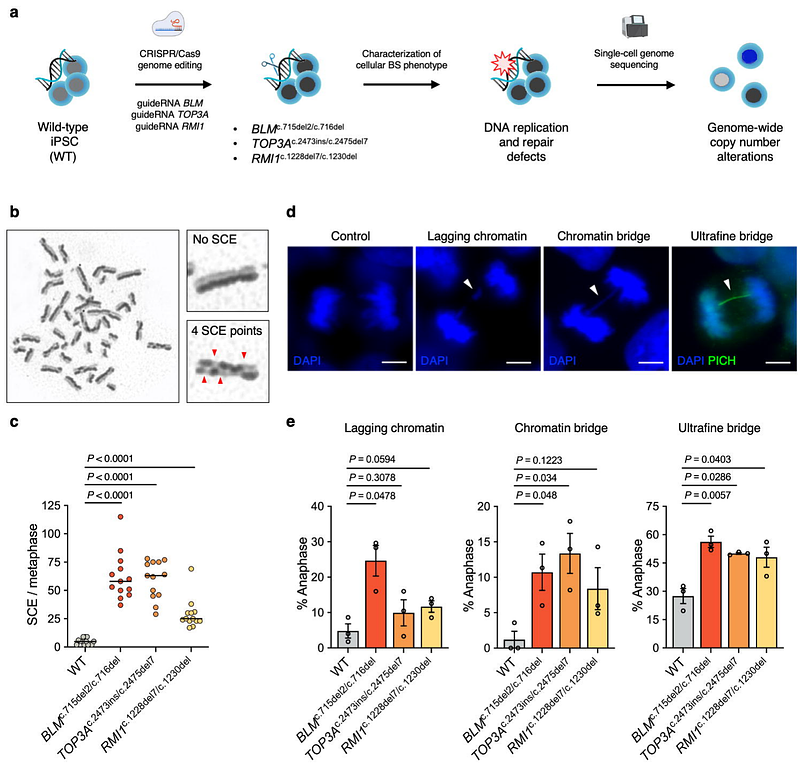
AbstractBiallelic loss-of-function (LoF) variants in the BTRR complex members BLM, TOP3A, RMI1, and RMI2 cause Bloom syndrome (BS). The BTRR complex mainly acts on DNA replication and DNA repair processes, and dysfunction of this complex underlies, e.g., increased genomic instability and cancer predisposition associated with the BS phenotype. Here, we report CRISPR/Cas9-based genome-edited isogenic induced pluripotent stem cell (iPSC) models with compound heterozygous LoF variants in BLM, TOP3A, and RMI1. The cellular phenotype of all three knockout (KO) iPSC lines included chromosome segregation defects, increased sister chromatid exchange rates, and impaired homologous recombination repair. Using single-cell whole genome sequencing, we showed that BTRR complex deficiency causes increased copy number alterations (CNAs) in the genomes and, therefore, represents a driver for genomic instability. CNA load was further induced by applying replication stress, and we observed that BTRRKO iPSCs acquired fewer de novo CNA events compared to wild-type cells, suggesting a possible limitation of genomic instability induction. Importantly, induced and non-induced CNAs in single-cell genomes were not stochastically distributed throughout the genome, but instead enriched at fragile sites. This finding might offer an opportunity for the development of novel NGS-based approaches to measure rates of genomic instability in disease conditions.