Functional Diversity in GII.4 Norovirus Entry: HBGA Binding and Capsid Clustering Dynamics
Functional Diversity in GII.4 Norovirus Entry: HBGA Binding and Capsid Clustering Dynamics
Ayyar, B. V.; Apostol, C. V.; Dave, J.; Kaundal, S.; Neill, F. H.; Ettayebi, K.; Maher, S.; Anish, R.; Larson, G.; Atmar, R. L.; Crawford, S. E.; Prasad, B. V. V.; Estes, M. K.
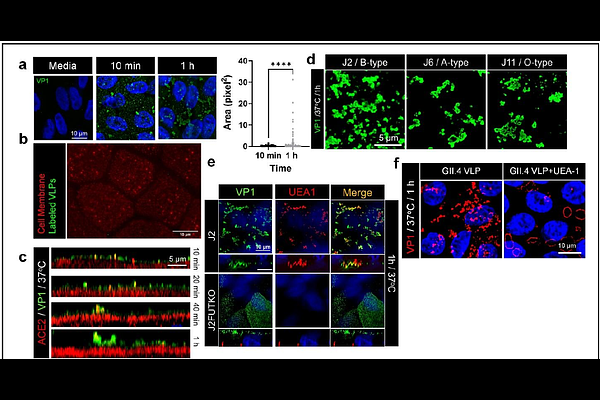
AbstractHuman noroviruses (HuNoVs), especially GII.4 strains, are the leading cause of acute viral gastroenteritis worldwide, yet no approved vaccines or antivirals exist. The pandemic GII.4 Sydney 2012 strain enters cells via membrane wounding and clathrin-independent carrier (CLIC)-mediated endocytosis, but it is unclear whether this entry mechanism is conserved across GII.4 variants. We compared early binding and entry of multiple GII.4 variants using wildtype and mutant GII.4 virus-like particles (VLPs) and modified human intestinal enteroid (HIE) cultures. Only a subset of GII.4 variants, including GII.4 Sydney, form distinct, HBGA-dependent capsid clusters on the cell surface. Clustering strains display significantly enhanced membrane wounding and endocytosis compared to non-clustering strains and outcompete non-clustering strains in replication assays as shown by complete inhibition of GII.4 Sydney replication. Using mutant VLPs and a HBGA non-binding mutant (R345A), we identified two residues, V333 and R339, in the VP1 protruding domain as critical mediators of clustering and entry. Mutations of these residues disrupt clustering and endocytosis without affecting HBGA binding, suggesting a role in post-attachment processes. While clustering and endocytosis are contingent upon VLP binding to HBGAs, inhibitor studies show they are independent of host protein glycosylation and are driven by lipid raft remodeling regulated by cholesterol and ceramides. Quantitative analyses across multiple GII.4 variants reveal an apparent dichotomy between clustering and non-clustering phenotypes, with clustering variants exhibiting higher entry competence. This distinction offers insight into strain-specific cell entry mechanisms and may aid in identifying the elusive proteinaceous HuNoV cellular receptor(s) supporting targeted therapeutic development.