Early-Life Environmental Exposures Reprogram Epigenomic Aging to Alter Gene Expression Trajectories
Early-Life Environmental Exposures Reprogram Epigenomic Aging to Alter Gene Expression Trajectories
Grimm, S. L.; Jangid, R.; Bartolomei, M. S.; Dolinoy, D. C.; Aylor, D. L.; Mutlu, G. M.; Biswal, S.; Zhang, B.; Wang, T.; TaRGET II Consortium, ; Coarfa, C.; Walker, C. L.
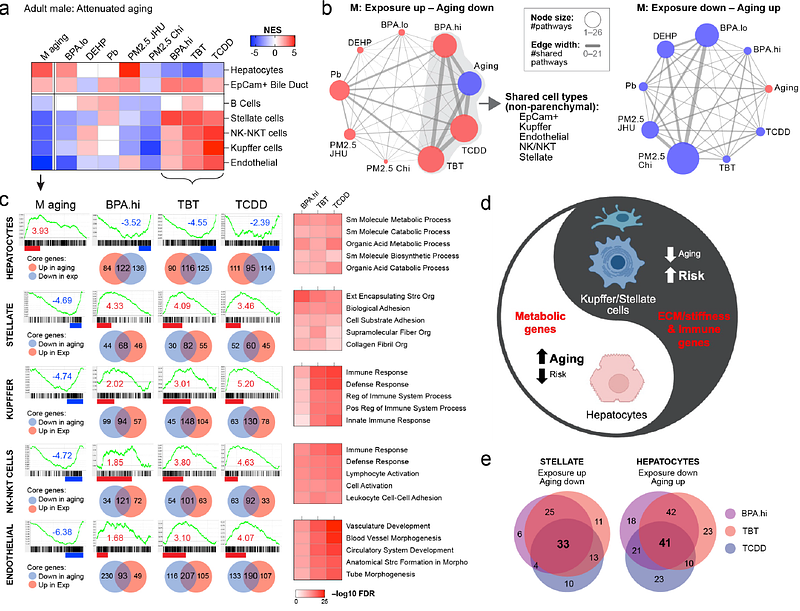
AbstractTo understand how early-life environmental exposures shape health and disease risk across the lifecourse, the TaRGET II Consortium exposed mice to diverse toxicants from pre-conception through weaning, and followed individual animals into adulthood, generating over 800 epigenomic and transcriptomic profiles. These profiles revealed that early-life exposures induced persistent epigenomic reprogramming and significantly disrupted the adult transcriptome. Notably, despite their diverse mechanisms of action, the exposure signatures of the xenoestrogen BPA, obesogen TBT, dioxin TCDD, and air pollutant PM2.5, were all largely comprised of genes normally differentially expressed during liver aging. Epigenetic histone modifications at enhancers - and, to a lesser extent, promoters - emerged as key targets for this reprogramming. Despite differing mechanisms of action, these four toxicants imparted similar \'fingerprints\' on the adult liver, characterized by direction- and cell type-specific polarization of the transcriptome. Hepatocyte genes that typically increase with age, particularly those in metabolic pathways, were downregulated, while conversely, non-parenchymal cell genes that typically decrease with age, such as those involved in extracellular matrix production, were upregulated. A similar signature of anti-correlation with programmed aging aging was also found in the transcriptome of patients with liver disease and hepatocellular carcinoma (HCC) and was effective at distinguishing healthy from diseased human livers. These findings demonstrate that the plasticity of epigenomic aging is vulnerable to early-life environmental exposures, which can reprogram the epigenome with lasting impacts on the transcriptome, and disease risk, later in life.