AI-Guided Discovery of LDHA Inhibitors Targeting Cancer Metabolism Using Machine Learning and Generative Chemistry: An End-to-End Drug Discovery Pipeline
AI-Guided Discovery of LDHA Inhibitors Targeting Cancer Metabolism Using Machine Learning and Generative Chemistry: An End-to-End Drug Discovery Pipeline
Petalcorin, M. F.; Petalcorin, M. I. R.
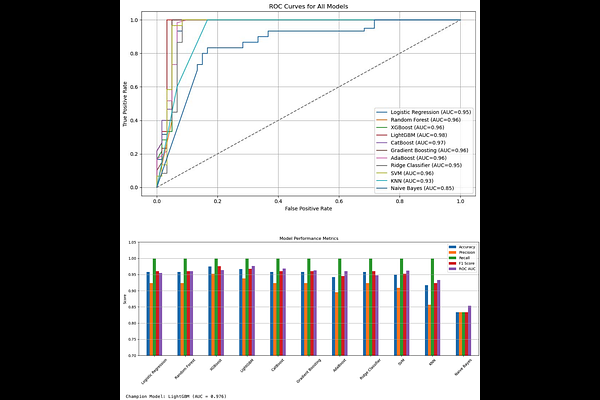
AbstractTargeting cancer metabolism has emerged as a promising therapeutic strategy, particularly through the inhibition of Lactate Dehydrogenase A (LDHA), a key enzyme that supports the Warburg effect in tumor cells. In this study, we present a comprehensive and fully reproducible machine learning (ML) and artificial intelligence (AI)-driven pipeline for the discovery of small-molecule LDHA inhibitors. By integrating bioactivity datasets from ChEMBL and BindingDB, along with natural products from COCONUT and AI-generated compounds from a ChemGPT-based molecular language model, we constructed a diverse and chemically rich screening library. Molecular descriptors were computed using Mordred, followed by feature selection, dataset balancing using SMOTE, and extensive model benchmarking across 11 classifiers. LightGBM was selected as the top-performing model with an AUC of 0.97. SHAP analysis provided model interpretability, revealing key molecular features influencing LDHA inhibition. Additionally, we trained ChemGPT on LDHA-specific SMILES in SELFIES format to generate 1,000 novel molecules, of which over 100 passed stringent drug-likeness, toxicity, and solubility filters. A subset exhibited high LDHA inhibition probabilities (>0.90) and structural novelty. This work highlights the potential of combining predictive modeling and generative chemistry for accelerating the early stages of cancer drug discovery and provides an open-source platform for continued development and validation.