Quantification of disease-associated RNA tandem repeats by nanopore sensing
Quantification of disease-associated RNA tandem repeats by nanopore sensing
Patino-Guillen, G.; Pesovic, J.; Panic, M.; Earle, M.; Ninkovic, A.; Petrusca, S.; Savic-Pavicevic, D.; Keyser, U. F.; Boskovic, F.
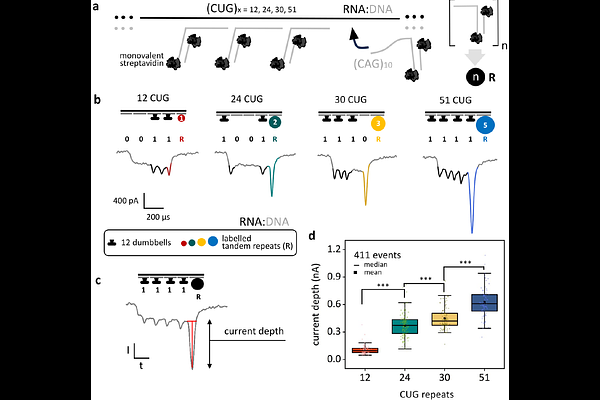
AbstractShort tandem repeat expansions underlie a class of neurological and neuromuscular diseases known as repeat expansion disorders (REDs), yet the precise characterization of these repeats remains technically challenging. Conventional amplification-based methods fail to resolve repeat length accurately due to amplification bias and sequence homogeneity. Here, we present a single-molecule nanopore-based strategy that enables direct quantification of tandem repeats in native RNA. By assembling RNA nanostructures that encode specific repeat number, we achieve repeat size discrimination with a resolution of 18 nucleotides. Using tandem repeat-containing RNA, we successfully detect and discriminate disease-relevant repeat lengths associated with myotonic dystrophy types 1 (DM1) and 2 (DM2), and congenital central hypoventilation syndrome-1 (CCHS1). Finally, we apply our method to total RNA extracted from a DM1 human cell line model, demonstrating its compatibility with complex biological samples. Our approach offers a platform for studying repeat expansion biology at the single-molecule level, with broad implications for diagnostics, clinical research and multiplexed repeat profiling.