A High-Dimensional Interfacial Wave Assay for Early Biophysical Profiling of Therapeutic Antibodies
A High-Dimensional Interfacial Wave Assay for Early Biophysical Profiling of Therapeutic Antibodies
St John, A. N.; Thomas, A. N.; Caramazza, P.; Meghdadi, A.; Bailey, J.; Lee, S.; Ramakrishna, V.; Amoruso, A. L. B.; Lam, E. S.-H.; Ganesh, N.; Shrivastava, S.
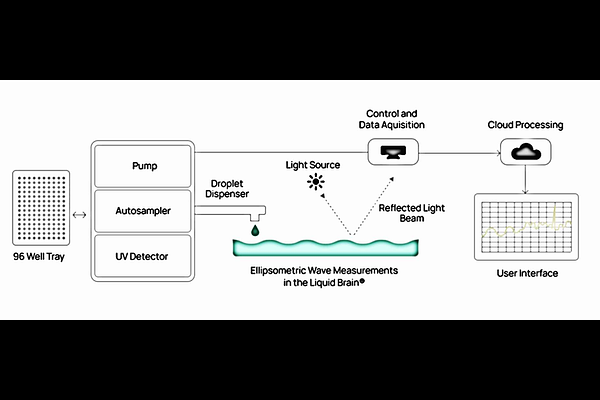
AbstractWe introduce the Variations of Interfacial Behaviour and its Evolution (VIBE) assay--a high-dimensional biophysical measurement inspired by the thermodynamics of biological signalling. In each assay, an antibody sample stimulates an excitable liquid interface held near a thermodynamic phase transition. The resulting response generates interfacial wave phenomena, including capillary, gravity, and Lucassen waves, encoded as dynamic surface signals using a proprietary technology stack, termed Liquid Brain(R). Each VIBE assay yields a rich biophysical fingerprint from minimal material ([≤]10 g), composed of descriptors derived from the recorded waveforms and their evolution. This paper makes two contributions: 1. Using a benchmark descriptor (VIBE1), we demonstrate that the assay identifies developability risk across a cohort of 236 clinical-stage antibodies. A single threshold ([≥]0.26) identifies 40 high-risk candidates with a 90.0% clinical failure rate, compared to 72.9% across the cohort, corresponding to a 1.23-fold enrichment for failure. Pre-excluding these outliers would have improved overall clinical success from 27.1% to 30.6% (~13% relative uplift). Importantly, VIBE1 is only weakly correlated with existing assays and provides orthogonal insight into failure risk, often flagging antibodies missed by standard panels. Correlation with in silico predicted structure-based descriptors in complementarity-determining regions (CDRs) suggests VIBE1 maybe driven by electrostatic and hydrophobic surface properties. 2. We establish that each assay yields multiple high-signal-to-noise (SNR) descriptors that are internally consistent, partially overlapping, and capture diverse aspects of biophysical behaviour. These form a stable, ML-ready biophysical fingerprint--suitable for predictive modelling, latent property inference, and mechanistic interpretation. While this paper stops short of training ML models, it defines the coordinate structure of this new descriptor space and makes a curated subset of features publicly available to enable independent evaluation. Together, these results position the VIBE assay as a minimal-material, multiplexed biophysical platform. The VIBE1 descriptor is ready for adoption by antibody developers as a conservative triage tool, while the broader descriptor landscape provides a new data layer for AI-native approaches to molecular profiling.