Deep homology and design of proteasome chaperone proteins in Candida auris
Deep homology and design of proteasome chaperone proteins in Candida auris
Rapala, J. R.; Siddiq, M.; Wittkopp, P. J.; O'Meara, M. J.; O'Meara, T. R.
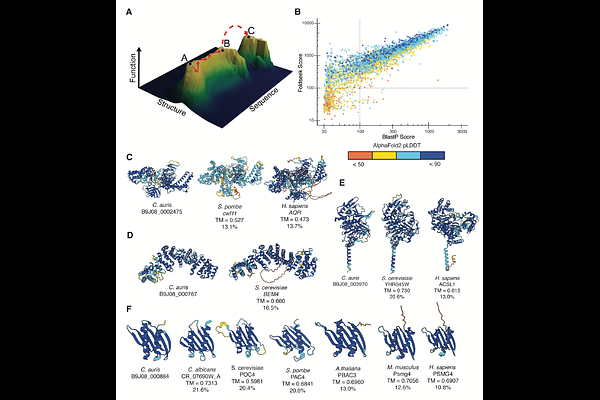
AbstractA central tenet of biology is that protein structure mediates the sequence-function relationship. Recently, there has been excitement about the promise of advances in protein structure modeling to generate hypotheses about sequence-structure-function relationships based on successes with controlled benchmarks. Here, we leverage structural similarity to identify rapidly evolving proteasome assembly chaperones and characterize their function in the emerging fungal pathogen Candida auris. Despite the large sequence divergence, we demonstrate conservation of structure and function across hundreds of millions of years of evolution, representing a case of rapid neutral evolution. Using the functional constraints on structure from these naturally evolved sequences, we prospectively designed de novo chaperones and demonstrate that these artificial proteins can rescue complex biological processes in the context of the whole cell.