DSB repair kinetics impair CRISPR-Cas9 editing in human embryos
DSB repair kinetics impair CRISPR-Cas9 editing in human embryos
Egli, D.; Marin, D.; Treff, N.; Jerabek, S.; Sung, J.; Xu, J.; Talukdar, J.
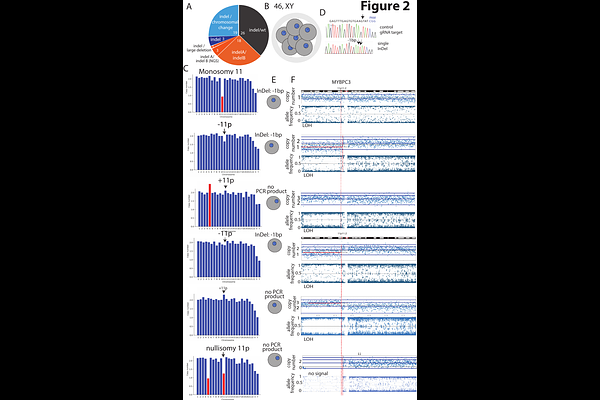
AbstractCas9 provides a powerful tool to interrogate DNA repair and to introduce targeted genetic modifications. However, a major challenge of Cas9-based editing in human embryos is the occurrence of chromosomal abnormalities caused by Cas9 cleavage. Furthermore, mosaicism - different genetic outcomes in different cells, prevent accurate genotyping using a single embryo biopsy. Through timed analysis of editing outcomes during the first cell cycle and timed inhibition of Cas9 using AcrIIA4, we show that most edits occur at least 12 hours post Cas9 injection and therefore after the first S-phase. This timing limits the ability to achieve uniform editing across cells. We found that segmental chromosomal abnormalities and the consequential loss of heterozygosity are common at Cas9 cleavage sites throughout the genome, including at MYBPC3 and at CCR5 loci, for which this has not previously been reported. Surprisingly, inhibiting Cas9 activity 8-12 hours before mitosis does not eliminate chromosomal aneuploidies. This suggests that double-strand break (DSB) repair in human embryos is exceedingly slow, with breaks remaining unrepaired for many hours. Thus, the timing of DSB induction and repair relative to the first S-phase and the first mitosis is intrinsically limiting to preventing mosaicism and maintaining genome stability in embryonic gene editing.