In silico design and validation of high-affinity RNA aptamers for SARS-CoV-2 comparable to neutralizing antibodies
In silico design and validation of high-affinity RNA aptamers for SARS-CoV-2 comparable to neutralizing antibodies
Yang, Y.; Qiao, L.; Jiang, Y.; Wang, Z.; Zhang, D.; Buratto, D.; Huang, L.; Zhou, R.
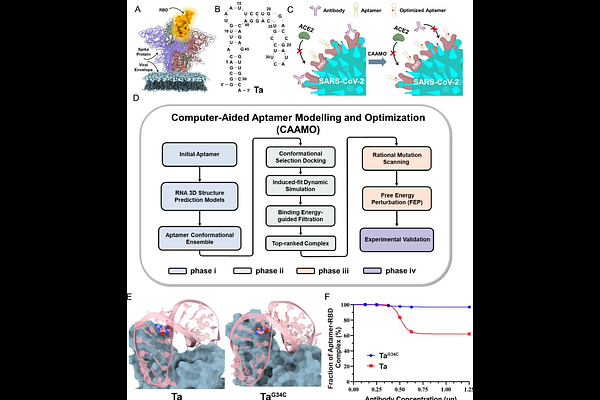
AbstractNucleic acid aptamers hold promise for clinical applications, yet understanding their molecular binding mechanisms to target proteins and efficiently optimizing their binding affinities remain challenging. Here, we present CAAMO (Computer-Aided Aptamer Modeling and Optimization), which integrates in silico aptamer design with experimental validation to accelerate the development of aptamer-based RNA therapeutics. Starting from the sequence information of a reported RNA aptamer, Ta, for the SARS-CoV-2 spike protein, our CAAMO method first determines its binding mode with the spike proteins receptor binding domain (RBD) through a multi-strategy computational approach. We then optimize its binding affinity via structure-based rational design. Among the six designed candidates, five were experimentally verified and exhibited enhanced binding affinities compared to the original Ta sequence. Furthermore, we directly compared the binding properties of the RNA aptamers to neutralizing antibodies, and found that the designed aptamer TaG34C demonstrated a comparable binding affinity to the RBD compared to all tested neutralizing antibodies. This highlights its potential as an alternative to existing COVID-19 antibodies. Our work provides a robust approach for the efficient design of a relatively large number of high-affinity aptamers with complicated topologies. This approach paves the way for the development of aptamer-based RNA diagnostics and therapeutics.