Dinucleotide codon mutations as a marker of diversifying selection in M. tuberculosis
Dinucleotide codon mutations as a marker of diversifying selection in M. tuberculosis
Zimenkov, D.; Ushtanit, A.
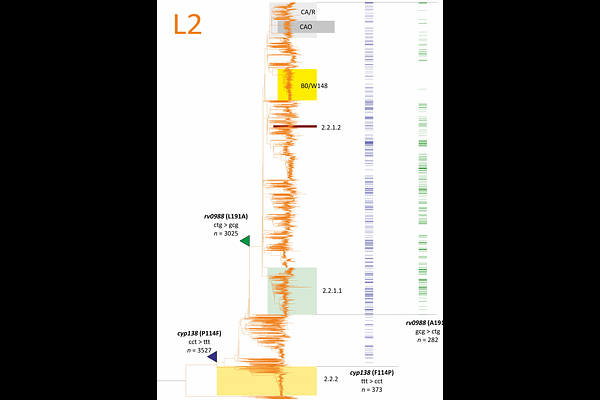
AbstractThe evolution of the human pathogen M. tuberculosis is shaped by various but interconnected processes of drug treatment pressure and host adaptation. We hypothesize that rarely accounted dinucleotide substitutions within a single codon, which allow for a broader range of amino acid substitutions than single nucleotide changes, are a significant aspect of diversifying selection. From the analysis of 43 studies, comprising 11,730 clinical isolates with resistance to rifampicin, 11 different dinucleotide substitutions were identified in 54 codons of resistance-determining regions of the rpoB gene. The prevalence of such substitutions is approaching 4%. Although rifampicin was introduced in treatment regimens in the 1970s, dinucleotide substitutions were also found in resistance determinants for newer drugs, linezolid and bedaquiline, rplC, and atpE, despite the significantly smaller number of resistant clinical isolates reported. Conducting a genome-wide analysis of dinucleotide mutations in the dataset of 9,941 genomes studied by the CRYpTIC Consortium, in addition to resistance determinants, we discovered three genes with a significantly elevated number of dinucleotide substitutions, which are presumably related to virulence and host adaptation. Two substitutions, cyp138 P114F and L191A are supposed to occur early in the evolutionary history of lineage 2 and are now under strong selection for reverse substitutions. Two amino acid substitutions in the third gene, rv2024c N508T and C514L, could also be obtained by single nucleotide changes and therefore are supposedly being selected based on frequency of codon usage. The signature of dinucleotide mutations introduces a novel approach to understanding the evolution of pathogen and identifying potential targets for antivirulence drugs. They underscore the complexity of the evolutionary dynamics within this pathogen, driven by diverse selection pressures, shedding light on the ongoing battle between M. tuberculosis and its human host.