Evolutionary dynamics of FoxQ2 transcription factors across metazoans: A tale of three ancient paralogs
Evolutionary dynamics of FoxQ2 transcription factors across metazoans: A tale of three ancient paralogs
Gattoni, G.; Lin, C.-Y.; York, J. R.; Keitley, D.; LaBonne, C.; Yu, J.-K.; Gillis, A.; Benito-Gutierrez, E.
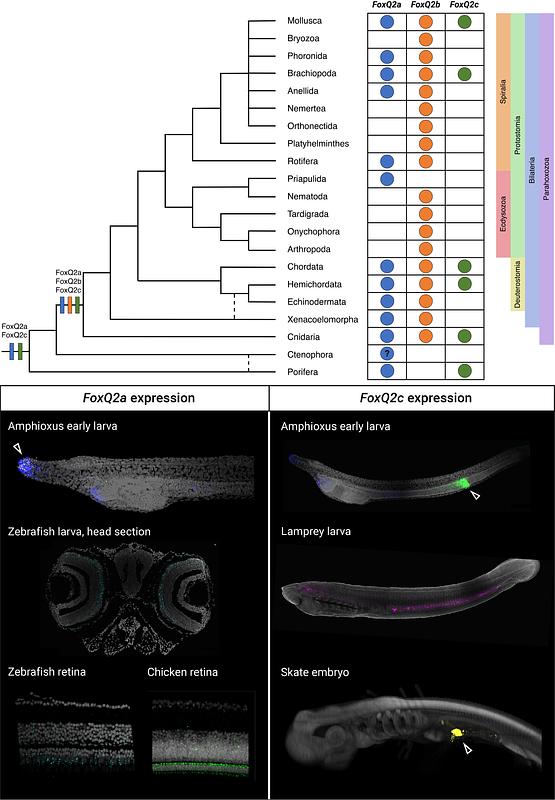
AbstractFoxQ2 is a highly conserved class of Forkhead-box transcription factors expressed on the anterior side of the body in cnidarians and bilaterians. Despite this conserved expression pattern, recent phylogenetic analyses have revealed a complex and rapid evolution of this class, with several taxon-specific duplications and losses. Until recently, FoxQ2 was thought to be lost in most vertebrate lineages, and its presence and localization in different vertebrate groups remains unclear. To reconcile these conflicting reports of conservation and divergence, here we present a comprehensive analysis of the phylogenetic relationships and expression patterns of FoxQ2 genes across metazoans. By combining phylogenetics and synteny analyses of FoxQ2 sequences from 21 animal phyla, we uncover the presence of three ancient FoxQ2 paralogs in bilaterians, which we name FoxQ2a, FoxQ2b and FoxQ2c. All three FoxQ2 paralogs are present in the chordate lineage and two are conserved in vertebrates, indicating a richer repertoire of vertebrate Fox genes than previously estimated. To investigate the expression of FoxQ2 genes across bilaterians, we mined expression data from existing single cell transcriptomic datasets of mollusk, acoel, amphioxus and zebrafish development, and expanded it using fluorescent in situ hybridization in amphioxus, lamprey, skate, zebrafish and chicken. Our analysis demonstrates the conserved anterior expression of FoxQ2a and FoxQ2b paralogs while also revealing a novel domain of FoxQ2c expression within the chordate endoderm, including in amphioxus, lamprey and skate. Finally, we devise a method to predict conserved transcription factor binding sites across the three extant amphioxus genera with specificity to developmental stage and cell-type identity. This suggests conserved regulatory interactions for the expression of FoxQ2a across deuterostomes. Overall, this work clarifies the complex evolutionary history of FoxQ2 genes and identifies a newly discovered endodermally-expressed Fox gene, FoxQ2c. We further propose that the early duplication of FoxQ2a and FoxQ2b, along with their redundant functions, provided the ideal background for subfunctionalization, contributing to the fast evolutionary rate of FoxQ2 sequences observed in bilaterians.