Bottom-up proteomics under acidic conditions using protease type XIII from Aspergillus saitoi
Bottom-up proteomics under acidic conditions using protease type XIII from Aspergillus saitoi
Tomioka, R.; Tomioka, A.; Ogata, K.; Ishihama, Y.
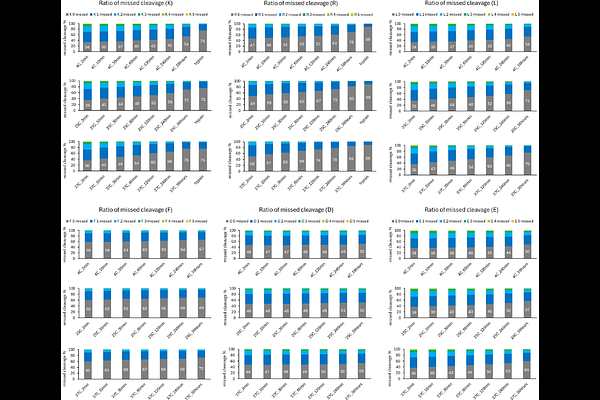
AbstractBottom-up proteomics is a powerful technique for comprehensive analysis of proteins by proteolytic cleavage followed by liquid chromatography/tandem mass spectrometry to identify the resulting peptides. Trypsin is the gold-standard protease for bottom-up proteomics, though its cleavage specificity limits peptide identification, depending on the protein sequence. In addition, its optimal pH is weakly alkaline, which can cause modification artifacts such as deamidation. We hypothesized that these limitations might be overcome by using protease type XIII (P13ase) from Aspergillus saitoi, which is active at low pH. P13ase has been used for protein structural analysis by hydrogen-deuterium exchange mass spectrometry, but its cleavage preferences have not been clarified. Here, we show that P13ase primarily cleaves the C-terminal side of Lys, Arg, and Leu, and the optimal P13ase digestion conditions for bottom-up proteomics are pH 3.5, 37C for 60 min. Under these conditions, sequence coverage of more than 90% was achieved for several proteins in HeLa cell extracts, which is unachievable with trypsin. In addition, P13ase digestion reduced artifacts such as deamidation products generated by cyclization reactions and subsequent hydrolysis. These results indicate that P13ase is a promising new tool for precision proteomics.