Conformational dynamics of the bacterial E3 ligase SspH1
Conformational dynamics of the bacterial E3 ligase SspH1
Kennedy, C. R.; Esposito, D.; House, D.; Rittinger, K.
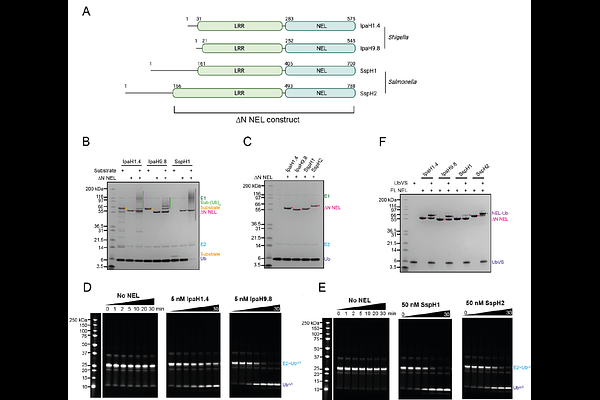
AbstractThe SspH/IpaH family of novel E3 ligases (NELs) are found in a number of Gram-negative bacteria and are used to target host enzymes for degradation to support pathogenesis. These E3 enzymes are autoinhibited in the absence of substrate and different models for release of autoinhibition have been suggested. However, many of the molecular details of individual steps during the ubiquitin transfer reaction remain unknown. Here, we present the crystal structure of Salmonella SspH1 and an analysis of the solution properties of SspH1 on its own and in complex with substrate and ubiquitin. Our data show that SspH1 exists in a conformational equilibrium between open and closed states and that substrate binding only modulates the distribution of these states but does not induce major conformational changes. This suggests that additional mechanisms must exist to bring the substrates close to the active site to mediate transfer of ubiquitin from the E3~Ub conjugate.