Thermodynamics and selection of the plasminogen activator inhibitor-1 latency transition
Thermodynamics and selection of the plasminogen activator inhibitor-1 latency transition
Haynes, L. M.; Holding, M. L.; Woodard, J.; Siemieniak, D.; Ginsburg, D.
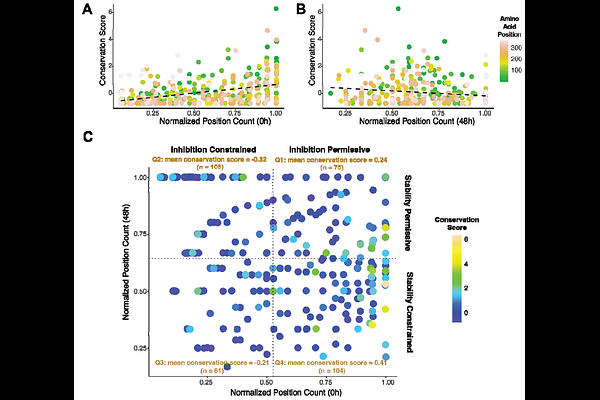
AbstractPlasminogen activator inhibitor-1 (PAI-1), like other serine protease inhibitors (SERPINs), exists in a functionally active metastable conformation. Unlike other SERPINs, PAI-1 undergoes a relatively rapid rearrangement to a lower energy latent conformation. We employ deep mutational scanning (DMS) to simultaneously probe the effects of nearly all single amino acid substitutions (92%) in PAI-1 on its latency transition as well as its ability to inhibit its canonical protease target, urokinase-type plasminogen activator (uPA). The DMS results are interpreted in the context of variant effect predictors (EVE, AlphaMissense, and CPT) and protein stability predictors (EvoEF and FoldX), as well as the evolutionary conservation of the PAI-1 sequence space across extant vertebrate species. We demonstrate that while variant effect predictors can generally partition PAI-1 as functional or non-functional with respect to uPA inhibition, they perform poorly when attempting to discriminate the effects of PAI-1 variants on its latency transition. However, by employing protein stability predictors, we demonstrate that the PAI-1 latency transition is most likely driven by changes in the energy barrier to the latency transition. Finally, we show that PAI-1's ability to inhibit uPA, as well as its thermodynamic instability in its active conformation are both under purifying selection that limits the sequence space available to PAI-1. These findings suggest that DMS of collateral fitness effects, including PAI-1's latency transition, may be better interpreted in contexts other than variant effect predictors, including protein thermodynamics and evolution.