Switching ON Hydrogen Sulfide: A Chemogenetic Toolkit for Spatially Resolved H2S Manipulation
Switching ON Hydrogen Sulfide: A Chemogenetic Toolkit for Spatially Resolved H2S Manipulation
Ghaffari Zaki, A.; Miri, S. M.; Vatandaslar, E.; Vilain, S.; Yigit, E. N.; Aydin, M. S.; Alp, M. I.; Eroglu, E.
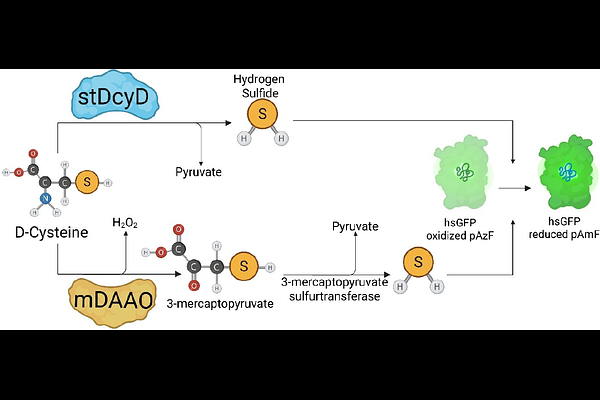
AbstractHydrogen sulfide (H2S) is emerging as a multifaceted signalling molecule that shapes energy metabolism, vascular tone, cancer biology and neurodegeneration. Although fluorescent and genetically encoded sensors now allow real-time H2S imaging, tools for selective control of intracellular H2S remain scarce. Here we benchmark two substrate-based chemogenetic enzymes - the yeast D-amino-acid oxidase (mDAAO) and the Salmonella typhimurium D-cysteine desulfhydrase (stDCyD) - for controlled H2S production. Both enzymes catalyse the conversion of D-cysteine to H2S, yet only mDAAO concomitantly generates hydrogen peroxide, introducing an unwanted oxidative signal. In contrast, stDCyD produces exclusively H2S, thereby offering a clean and efficient means to elevate intracellular H2S in vitro. Our data position stDCyD as the superior chemogenetic actuator for dissecting H2S biology without confounding redox artefacts.