Quantitative analysis of inhibitor-induced assembly disruption in human UDP-GlcNAc 2-epimerase using mass photometry
Quantitative analysis of inhibitor-induced assembly disruption in human UDP-GlcNAc 2-epimerase using mass photometry
Boback, N.; Gorenflos Lopez, J. L.; Hackenberger, C. P.; Di Lella, S.; Lauster, D. C.
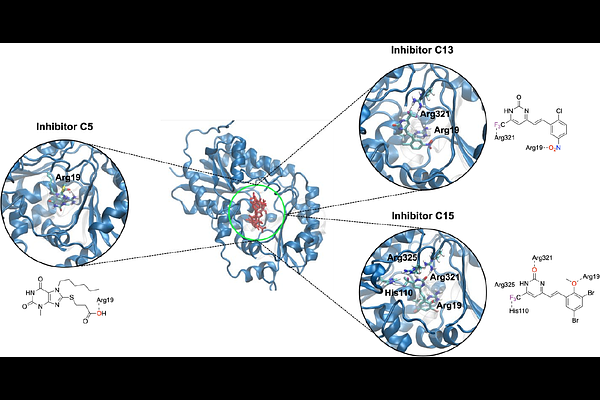
AbstractThe enzyme UDP-GlcNAc 2-epimerase/N-acetylmannosamine kinase (GNE/MNK) serves as the rate-limiting catalyst in the de novo biosynthesis of sialic acid, a molecule essential for cellular signaling, adhesion, and immunity. Modulating GNE activity is a potential therapeutic strategy for many diseases. Building on our previous finding that GNE activity is coupled to its tetramerization, we sought for a deeper understanding of GNE assembly equilibria in the presence of the substrate UDP-GlcNAc, as well as of the inhibitors C5, C13, and C15. While substrate addition enhanced dimer-dimer affinity of GNE by 120-fold and reduced monomer-monomer affinity by 5-fold, the inhibitors demonstrated dose-dependent inhibitory effects on tetramer stability, with IC50 values in the low micromolar range. Molecular docking studies revealed binding energies that modulate allostery in dimer and tetramer formation. These findings advance the mechanistic understanding of GNE in sialic acid biosynthesis and highlight iSCAM as a powerful tool for analyzing protein assembly and inhibition.