High-Throughput Machine Learning-Aided Antibody Discovery for Cell Surface Antigens
High-Throughput Machine Learning-Aided Antibody Discovery for Cell Surface Antigens
Kothiwal, D.; Kollasch, A. W.; Hollmer, N.; Ghosh, A.; Zhang, R.; Anuganti, M.; Paul, S. B.; Zagar, Y.; Abdollahi, M.; Anderson, Z.; Belay, F.; Salotto, M.; Ulmer, S.; AbdelAlim, Y. A.; Kumar, S.; Vangala, M.; Yang, C.; Chedotal, A.; Jardine, J. G.; Teixeira, A. A. R.; Moshinsky, D. J.; Zhu, H.; Zhu, S.; Springer, T. A.; Marks, D. S.; Meijers, R.
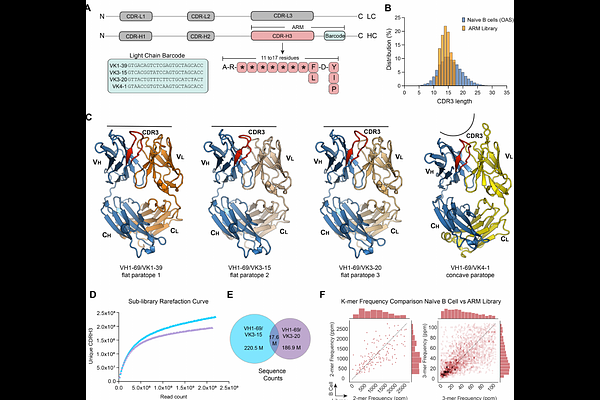
AbstractMachine learning (ML) has the potential to revolutionize antibody design and selection, but its success depends on access to extensive, well-curated datasets of antibody-antigen interactions. To address this need, we developed a synthetic Fab yeast display library optimized for seamless ML integration, focusing on sequence diversity within the CDRH3 loop. The library incorporates key sequence features derived from human B cell repertoires essential for efficient antibody generation captured in a compact antigen recognition module (ARM) format. Built using the VH1-69 heavy chain and four light chains, the library was evaluated against ten human and murine cell surface antigens, including PD-L1, TIGIT, and ROBO1. This approach yielded hundreds of antibodies with robust biophysical properties, validated for functional performance in flow cytometry and immunohistochemistry. Furthermore, ML analysis identified additional antibodies for ROBO2 and PD-L2 from the aggregate sequencing data, demonstrating utility for hybrid in silico and experimental workflows. We provide a publicly accessible dataset comprising more than 68,000 Fab sequences and 486 characterized antibodies. This study establishes an ML-compatible framework designed to accelerate and streamline antibody discovery and development.