Liposome Encapsulation Enables Near-Native Cryo-EM Structural Determination by Shielding Macromolecules from Non-Physiological Interfaces
Liposome Encapsulation Enables Near-Native Cryo-EM Structural Determination by Shielding Macromolecules from Non-Physiological Interfaces
Zikai, G.; Lijuan, M.; Hang, F.; Chenguang, Y.; Dongfei, M.; Hongtao, Z.; Yumei, W.; Dapeng, S.; Shuxin, H.; Chunhua, X.; Wei, D.; Jianbing, M.; Ying, L.; Ming, L.
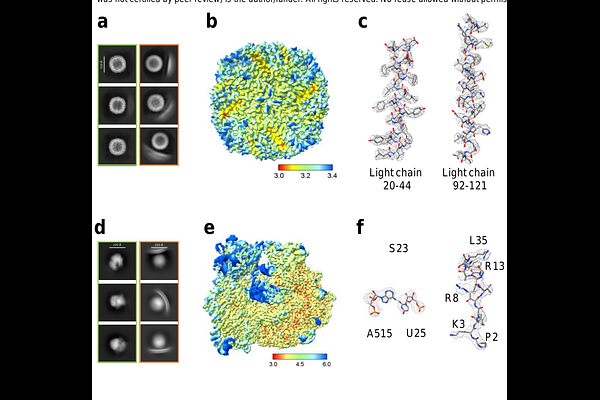
AbstractCryo-electron microscopy (cryo-EM) has emerged as a very powerful tool for high-resolution structure determination of macromolecule complexes. However, sample preparation remains a major bottleneck in cryo-EM workflows. Most structures were resolved not in a native solution environment, but rather in the non-physiological interfaces such as the air-water interface (AWI), where the folding/unfolding energy landscape of macromolecules may be significantly altered, leading to artifact, preferential orientation, particle disassembly or even denaturation. To address this challenge, we developed a robust, sample-independent method utilizing liposome encapsulation that preserve macromolecules in a solution-like and near native environment throughout sample preparation. Using equine spleen apoferritin and the Escherichia coli (E. coli) ribosome as model systems, we demonstrate efficient particle incorporation into liposomes and successful high-resolution structure determination. Notably, our method yields significantly reduced particle disassembly, denaturation, and preferential orientation compared to samples on holey carbon and graphene grids. We anticipate that this approach will facilitate structural studies of other challenging macromolecular complexes that are sensitive to interfacial effects.