Evolutionary trajectories of cohesin gatekeepers across the Tree of Life
Evolutionary trajectories of cohesin gatekeepers across the Tree of Life
Quiroz, D.; Pierce, A.; Monroe, J. G.
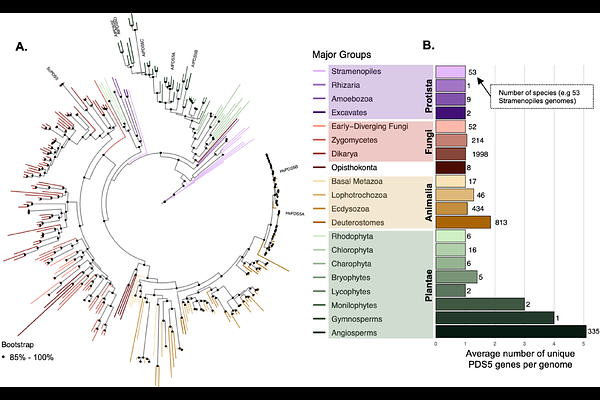
AbstractEukaryotic genomes are physically organized into chromatin which regulates virtually every dimension of genome biology including replication, transcription, and DNA repair. Precocious Dissociation of Sisters 5 (PDS5) protein is a key regulator of the cohesin complex, responsible for generating higher order chromatin structures within and between chromosomes. In yeast and vertebrates PDS5 can lead to cohesin establishment or disassembly, depending on its interaction with other proteins. In this study, we explored the diversity of PDS5 proteins across eukaryotes; HEAT-repeats forming a hook-like structure (PDS5 domain), followed by a predicted disordered region of variable lengths, are observed across the Tree of Life. Interestingly, two homolog types (A and B) with signs of varying chromatin localization emerged independently in later branches of animal and plant kingdoms, in vertebrates and vascular plants, respectively (~400-500 MYA). We also investigated the emergence of two plant-specific features in the PDS5 proteins: 1.) An ancestral acquisition of a Tudor domain with conserved H3K4me1-binding amino acids in land plants 2.) A truncated version of PDS5 proteins that lacks essential features of PDS5 domain and is involved in the RNA-directed DNA methylation (RdDM) plant-specific pathway which emerged in seed plants. This work provides the most comprehensive study of PDS5 diversity, and suggests the emergence of specialized cohesin regulatory mechanisms that could shape distinct strategies for genome organization across eukaryotes.