Comprehensive molecular impact mapping of common and rare variants at GWAS loci
Comprehensive molecular impact mapping of common and rare variants at GWAS loci
Balderson, B.; Tule, S.; Okino, M.-L.; Rieger, W. J.; Corban, S.; Jaureguy, J.; Palpant, N.; Gaulton, K. J.; Boden, M.; McVicker, G.
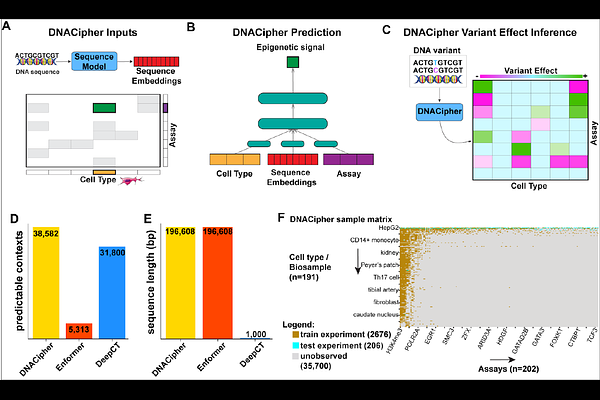
AbstractDeep learning sequence to function models can predict the molecular effects of genetic variants, but their predictions are limited to the cell types and assays they are trained on. Here we describe DNACipher, a deep learning model that predicts the effects of genetic variants across diverse biological contexts-including those not directly measured. DNACipher takes 196 kb of genome sequences as input and imputes variant effects across 38,582 cell type-assay combinations. DNACipher generates predictions for >7 times as many contexts as Enformer, which allows for better detection of variant effects at expression quantitative trait loci (eQTLs). We also introduce DNACipher Deep Variant Impact Mapping (DVIM), a method to identify variants with molecular effects at genome-wide association study (GWAS) loci. Application of DVIM to type 1 diabetes (T1D) reduced the mean fine-mapping credible set size from 24 to 1.4 variants per signal. DVIM variants had significantly higher fine-mapping posterior probabilities, and their predicted effects were supported by single-nucleus ATAC-seq and luciferase assays. DVIM also detected 6547 rare variants with molecular effects at 96% of GWAS T1D loci, and these were enriched for associations with immune traits. In summary, DNACipher DVIM prioritises common and rare variants at GWAS loci by predicting molecular effects across a broad range of contexts.