Untangling the effects of within-host and between-host natural selection in viruses with applications to evolution of HIV-1 co-receptor usage
Untangling the effects of within-host and between-host natural selection in viruses with applications to evolution of HIV-1 co-receptor usage
Drake, K.; Franceschi, V. B.; Nascimento, F. F.; Volz, E. M.
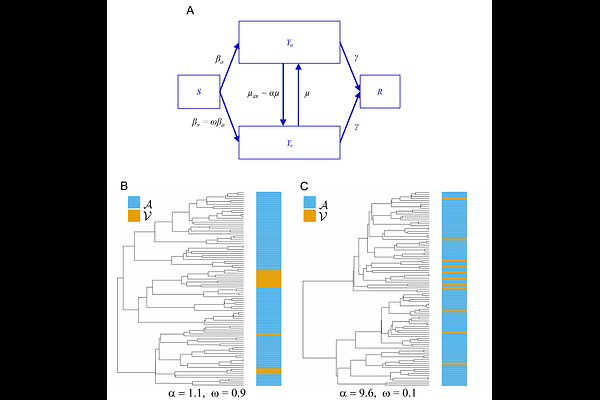
AbstractThe fitness of a pathogen can be characterised at multiple levels corresponding to different stages of its life cycle, including replication within target cells (replicative capacity), immune evasion, and the capacity to infect new hosts (transmissibility). Pathogens evolve towards greater fitness within hosts, which can potentially compromise transmissibility. Typically, epidemiological studies for measuring pathogen fitness compute a single selection coefficient for pathogen variants that encompasses all stages of the pathogen life cycle. To address this problem, we develop a population-genetic model and software for the inference of multi-level selection coefficients which represent relative within-host fitness and transmissibility. Our approach builds on population genetic models previously applied to macro-evolution, whereby speciation and extinction rates are state-dependent. We relate the state-dependence in speciation and extinction rates to competing within- and between-host selection. Evolution of beneficial phenotypes within hosts may occur rapidly (higher 'speciation'), but reduce rates of dispersal between hosts (higher 'extinction'). This new coalescent-based method also accounts for highly nonlinear-population dynamics and variable sampling through time making it suitable for epidemiological studies. We derive the equilibrium proportion of a variant with multi-level selection under mutation-selection balance and show how this can be used in combination with the coalescent to greatly increase accuracy and precision of estimated selection coefficients. We characterise the accuracy of this method using a variety of birth-death and coalescent simulations before analysing a real dataset of HIV-1 genetic sequences from a large population-based sample of 4056 people living with HIV in Zambia. We quantified the multi-level relative fitness underlying HIV-1 'co-receptor switching'. In accordance with previous observations, we estimate that the relative transmissibility of X4-tropic virus is 9%, while within-host switching to X4-tropism is approximately seven-fold higher than switching to R5-tropism, indicating a strong within-host and between-host tradeoff of co-receptor usage. Our methods are available in an open-source R package musseco, providing a general-purpose tool for inferring multi-level selection coefficients for epidemiological sequence data.