Subcellular Mass Spectrometry Reveals Proteome Remodeling in an Asymmetrically Dividing (Frog) Embryonic Stem Cell
Subcellular Mass Spectrometry Reveals Proteome Remodeling in an Asymmetrically Dividing (Frog) Embryonic Stem Cell
Shen, B.; Pade, L. R.; Zhou, F.; Nemes, P.
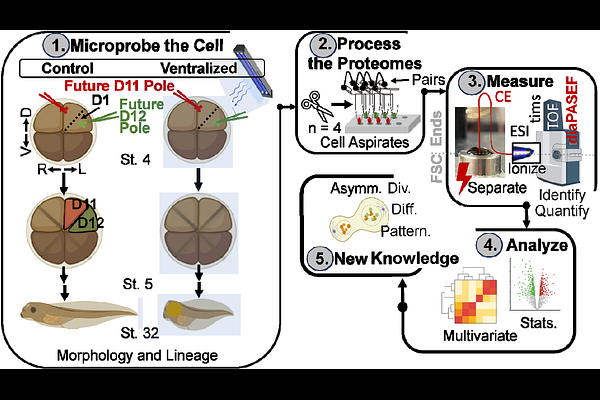
AbstractSubcellular proteomics holds the potential to reveal the molecular architecture of cellular processes with unprecedented spatial resolution. Performing these analyses deeply, at the level of hundreds to thousands of proteins in subcellular resolution, is a high and still unmet technical need. Here, we advance microprobe capillary electrophoresis mass spectrometry (CE-MS) to achieve deep proteomic coverage, quantifying over 1,000 proteins within opposing poles of an asymmetrically dividing embryonic stem cell (blastomere). We integrated CE electrospray ionization (CE-ESI) with trapped ion mobility spectrometry time-of-flight (timsTOF) MS, implementing data-independent acquisition (DIA) via parallel accumulation serial fragmentation (diaPASEF). This CE-diaPASEF workflow identified 1,035 proteins from ca. 200 pg of proteome digest, equivalent to ca. 80% contnet of a HeLa cell, with high reproducibility (coefficient of variation less than 15% across technical triplicates). With microprobe sampling, this technology quantified 808 to 1,022 proteins in opposing poles of a dorsal-animal (D1) blastomere in the 8-cell Xenopus laevis embryo. Comparative proteomic analysis of the D1 blastomere and its descendants, the dorsal-animal-midline (D11) and dorsal-animal-lateral (D12) cells, revealed diverse molecular outcomes of asymmetric division: some protein profiles remained conserved, while others underwent significant or even reversed changes as these lineages descended into neural tissue and epidermal trajectories. Ultraviolet light-induced ventralization was performed to help disentangle subcellular gradients from dorsal-ventral patterning. Collectively, this work establishes microprobe CE-diaPASEF as a powerful platform for deep subcellular proteomics, enabling new insights into spatial proteome organization during key developmental processes.