Focus on single gene effects limits discovery and interpretation of complex trait-associated variants
Focus on single gene effects limits discovery and interpretation of complex trait-associated variants
Lawrence, K. A.; Gjorgjieva, T.; Montgomery, S. B.
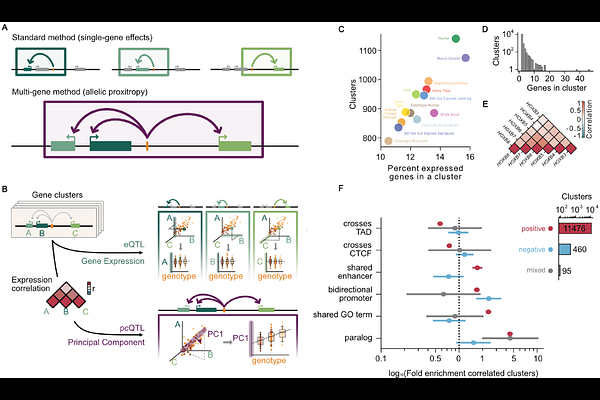
AbstractStandard QTL mapping approaches consider variant effects on a single gene at a time, despite abundant evidence for allelic pleiotropy, where a single variant can affect multiple genes simultaneously. While allelic pleiotropy describes variant effects on both local and distal genes or a mixture of molecular effects on a single gene, here we specifically investigate allelic expression \"proxitropy\": where a single variant influences the expression of multiple, neighboring genes. We introduce a multi-gene eQTL mapping framework - cis-principal component expression QTL (cis-pc eQTL or pcQTL) - to identify variants associated with shared axes of expression variation across a cluster of neighboring genes. We perform pcQTL mapping in 13 GTEx human tissues and discover novel loci undetected by single-gene approaches. In total, we identify an average of 1396 pcQTLs/tissue, 27% of which were not discovered by single-gene methods. These novel pcQTL colocalized with an additional 142 GWAS trait-associated variants and increased the number of colocalizations by 34% over single-gene QTL mapping. These findings highlight that moving beyond single-gene-at-a-time approaches toward multi-gene methods can offer a more comprehensive view of gene regulation and complex trait-associated variation.