Conserved spatial patterning of gene expression in independent lineages of C4 plants
Conserved spatial patterning of gene expression in independent lineages of C4 plants
Sun, T.; Bonthala, V. S.; Stich, B.; Luginbuehl, L. H.; Hibberd, J. M.
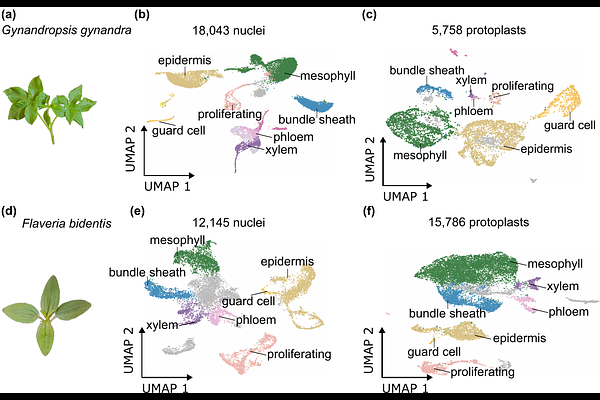
AbstractC4 photosynthesis enhances carbon fixation efficiency by reducing photorespiration through the use of an oxygen-insensitive carboxylase and spatial separation of photosynthesis between mesophyll and bundle sheath cells. The C4 pathway has evolved independently in more than sixty plant lineages but molecular mechanisms underpinning this convergence remain unclear. To explore this, we generated high-resolution transcriptome atlases for two independently evolved C4 dicotyledonous species - Gynandropsis gynandra (NAD-ME subtype) and Flaveria bidentis (NADP-ME subtype). We used both single-cell and single-nucleus RNA sequencing to capture gene expression profiles from individual leaf cells, enabling detailed comparison of cell types and transcriptional signatures. Both approaches produced biologically comparable data for major leaf cell types, transcriptomes from single-nucleus sequencing showed lower stress signatures and were more representative of tissue proportions in the leaf. The single-nucleus data revealed that bundle sheath cells from both C4 species share a gene expression pattern associated with mesophyll cells of C3 plants. A conserved set of transcription factors, including members of the C2H2 and DOF families, was identified in the bundle sheath cells of both species. This study presents the first single-cell-resolution transcriptomes for two independent C4 dicot lineages and provides a valuable resource, including a web-based portal for data visualization.