Membranous interacting partners of phage-type plastid RNA polymerase have limited impact on plastid gene expression during chloroplast development
Membranous interacting partners of phage-type plastid RNA polymerase have limited impact on plastid gene expression during chloroplast development
Kurotaki, Y.; Nishino, A. S.; Fujii, S.
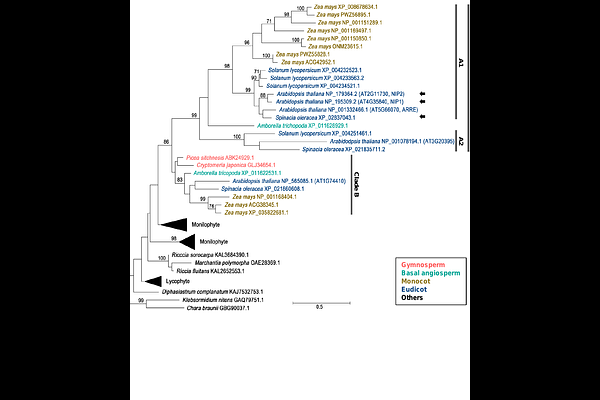
AbstractIn vascular plants, genes in the plastid genome are transcribed by two types of RNA polymerases, namely bacterial-type nuclear-encoded and phage-type plastid-encoded plastid RNA polymerases (NEP and PEP, respectively). Eudicots, including Arabidopsis, carry two isoforms of NEP, RPOTp and RPOTmp. NEPs transcribe multiple plastid-encoded genes including subunits of PEP and translocon, thus are indispensable for the maintenance of plastids. However, regulatory mechanisms of NEPs are largely unknown. RPOTmp transcribes the 16S rRNA gene from a specific promoter in the seeds during vernalization, and its mutation in Arabidopsis retards the chloroplast development. As interacting partners of RPOTmp, two NEP-INTERACTING PROTEINs (NIP1 and NIP2) have been identified and suggested to suppress RPOTmp activity by tethering RPOTmp to the thylakoid membrane during chloroplast development in the presence of light, but their precise roles in transcriptional regulation remain to be addressed. From these previous reports, we hypothesize that functions of RPOTmp would depend on the light conditions and expression of NIPs. To gain insight into how RPOTmp is controlled, we performed functional analysis of RPOTmp and NIPs using Arabidopsis mutants during germination in the dark and de-etiolation processes under light. We found that RPOTmp-dependent transcription of 16S rRNA is active in imbibed seeds and remains basal level throughout the post-germination processes, regardless of light conditions. We also demonstrated a limited impact of NIPs on RPOTmp function during these processes. Our phylogenetic analysis indicates that NIPs have distinct evolutionary profiles compared to RPOTmp, and Arabidopsis is unlikely to have additional NIP-like proteins in plastids. Based on these findings, we propose a modified model of RPOTmp regulation during chloroplast development: RPOTmp activity remains stable throughout the process of the chloroplast differentiation and is unaffected by light and NIPs.