VN1K: a genome graph-based and function-driven multi-omics and phenomics resource for the Vietnamese population
VN1K: a genome graph-based and function-driven multi-omics and phenomics resource for the Vietnamese population
Tran, T. T. H.; Hoang, T. H.; Tran, M. H.; Nguyen, N. T.; Nguyen, D. T.; Pham, T.; Pham, T. M.; Nguyen, N. N.; Vu, G. M.; Duong, V. C.; Vu, Q. T.; Nguyen, T. K.; Nguyen, T. M.; Vu, H. Q.; Dang, T.; Nguyen, H.; Do, T.; Le, C.; Nguyen, H. T. T.; Le, N. Q.; Le, L. T.; Vu, D. M.; Ngo, T. D.; Le, H. T. T.; Nguyen, L. T.; Ha, T. C.; Hoang, Y.; Dao, D. X.; Giang, P. H.; Luu, H. N.; Dao, M. D.; Le, L.; Le, V. S.; Tran, T.; Nguyen, Q.; Le, D.-H.; Nguyen, D. T.; Vu, V. H.; Vo, N. S.
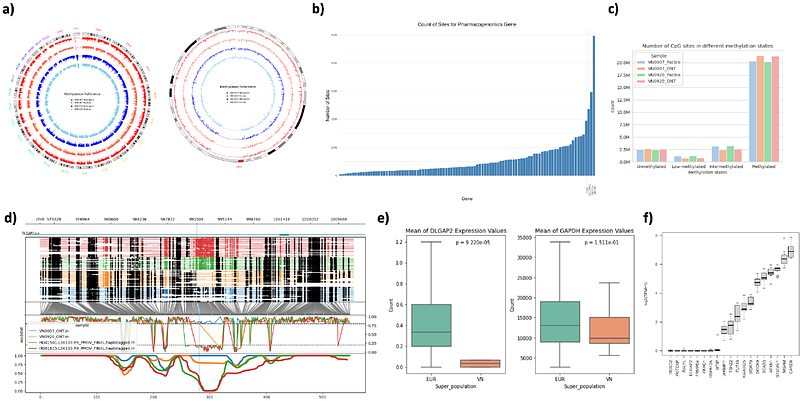
AbstractVietnam, the 16th most populated nation, remains profoundly underrepresented in global genomic databases. Here, we present VN1K, a first-ever comprehensive and well-curated resource of multi-omics data with a wide-range of phenotypic information of 1,011 unrelated Vietnamese individuals. High-depth short-read whole-genome sequencing data were generated for all samples along with various -omic data, including microarray, long-read whole-genome sequencing, and RNA sequencing. Using a high-sensitivity variant detection pipeline, which included a pangenome graph reference and a deep-learning framework, we identified nearly 40 million variants of which 8.5 million are novel with nearly 900 thousand short insertions/deletions and 39 thousand structural variants. Specifically, VN1K featured a first-ever whole-genome methylation profile based on long read sequencing. A genotype imputation panel was also created with the highest accuracy on the Vietnamese population. Variants with significantly different allele frequencies in the Vietnamese population compared to others were found to be functionally significant, especially in genes associated with immune diseases (HLA-B, KIR3DL3, KIR2DL1, KIR2DL4) or drug responses (CYP2C19, CYP2D6, VKORC1, CYP2B6). We were also able to map various loci related to hepatitis B virus infection as well as six disease traits, including triglyceride levels, LDL-C, serum glucose levels, HbA1c, and levels of two liver enzymes (ALT and AST). VN1K dataset is accessible via genome.vinbigdata.org, an integrated platform with both linear and graph-based genome browser for facilitating data exploration, research, and applications in precision medicine.