Simulation-based spatially explicit close-kin mark-recapture
Simulation-based spatially explicit close-kin mark-recapture
Patterson, G.; Goodfellow, C. K.; Ting, N.; Kern, A. D.; Ralph, P. L.
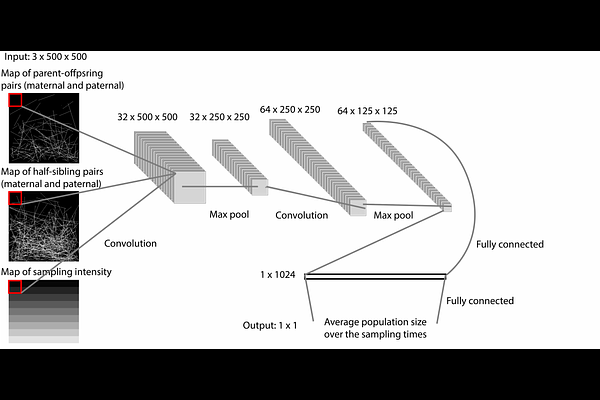
AbstractEstimating the size of wild populations is a critical priority for ecologists and conservation biologists, but tools to do so are often labor intensive and expensive. A promising set of newer approaches are based on genetic data, which can be cheaper to obtain and less invasive than information from more direct observation. One of these approaches is close-kin mark-recapture (CKMR), a type of method that uses genetic data to identify kin pairs and estimates population size from these pairs. Although CKMR methods are promising, a major limitation to using them more broadly is a lack of CKMR models that can deal with spatial heterogeneity both in population density and sample effort. We introduce a simulation-based approach to CKMR that uses spatially explicit individual-based simulation in concert with a deep convolutional neural network to estimate population sizes. Using extensive simulation, we show that our method, CKMRnn, is highly accurate, even in the face of spatial heterogeneity, and demonstrate that it can account for potential confounders such as unknown population histories. Finally, to demonstrate the accuracy of our method in an empirical system, we apply CKMRnn to data from a Ugandan elephant population, and show that point estimates from our method recapitulate those from traditional estimators but that the confidence interval on our estimator is reduced by approximately 30%.