H3K79 methylation and H3K36 tri-methylation synergistically regulate gene expression in pluripotent stem cells
H3K79 methylation and H3K36 tri-methylation synergistically regulate gene expression in pluripotent stem cells
Cooke, E. W.; Zeng, C.; Nur, S. M.; Jia, Y.; Huang, A.; Chen, J.; Gao, P.; CHEN, F. X.; Jin, F.; Cao, K.
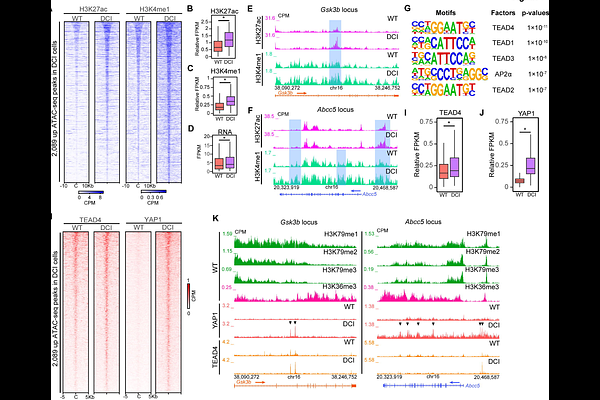
AbstractIn metazoans, nucleosomes harboring H3K79 methylation (H3K79me) deposited by the histone methyltransferase DOT1L decorate actively transcribed genes. Although DOT1L is implicated in transcription regulation and pathogenesis of human diseases such as leukemia and neurological disorders, the role of H3K79me in these biological processes remains elusive. Here, we reveal a novel functional synergism between H3K79me and H3K36 tri-methylation (H3K36me3), another histone modification enriched at active genes, in regulating gene expression and neural cell fate transition. Simultaneous catalytic inactivation of DOT1L and the H3K36 methyltransferase SETD2 via gene editing leads to the global loss of H3K79me and H3K36me3, hyperactive transcription, and failures in neural differentiation. Interestingly, the loss of H3K79me and H3K36me3 causes increased transcription elongation, gained chromatin accessibility at a group of enhancers, and increased binding of TEAD4 transcription factor and its co-activator YAP1 at these enhancers. Furthermore, YAP-TEAD inhibition partially restores the expression levels of hyperactivated genes upon H3K79me/H3K36me3 loss. Taken together, our study demonstrates a synergistic role of H3K79me and H3K36me3 in regulating transcription and cell fate transition, unveils novel mechanisms underlying such synergism, and provides insight into designing therapies that target diseases driven by misregulation or mutations of DOT1L and/or SETD2.