High-throughput engineering and modification of non-ribosomal peptide synthetases based on Golden Gate assembly
High-throughput engineering and modification of non-ribosomal peptide synthetases based on Golden Gate assembly
Podolski, A.; Lindeboom, T. A.; Praeve, L.; Kranz, J.; Schindler, D.; Bode, H. B.
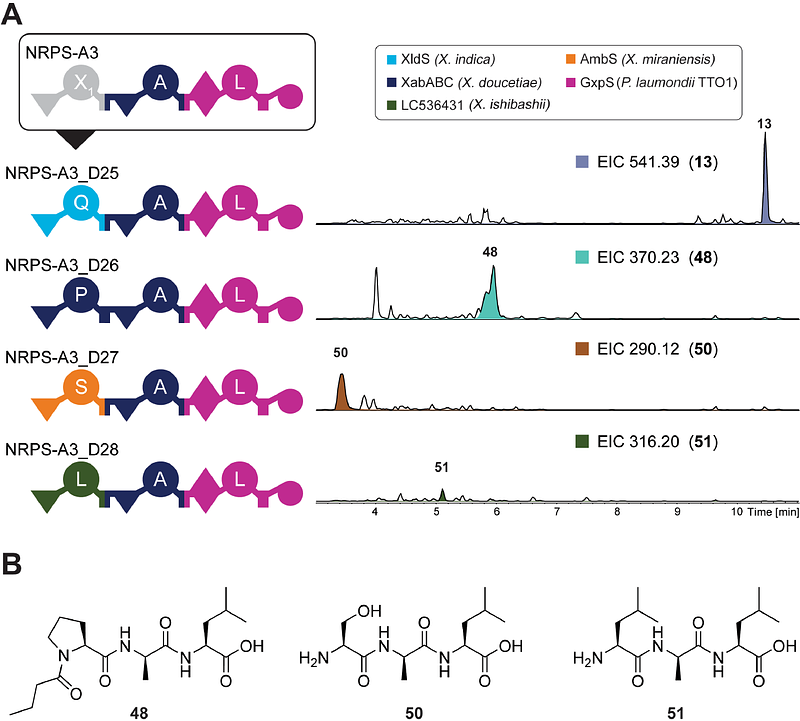
AbstractNon-ribosomal peptide synthetases (NRPS) are multimodular enzymes that produce complex peptides with diverse biological activities, potentially being used as clinical drugs. However, the pharmaceutical applications of such natural peptides often require further derivatisation and modification of the peptide backbone, mainly performed by chemical synthesis. A sustainable alternative resembles the in vivo engineering of NRPS to change and modify the enzyme properties rationally and, thus, the produced products. The novel NRPS engineering concept, the eXchange Unit Thiolation domain (XUT), allows the efficient modular assembly of different natural NRPS fragments to form hybrid NRPS that produce defined peptides. In this study, we describe a Golden Gate assembly (GGA) method for efficient high-throughput generation of novel and engineered NRPS libraries utilising the XUT concept. This method was applied to generate over 100 novel NRPS with the possibility of changing starter, elongation, and termination modules, respectively. Additionally, we applied this method for targeted modification of the xenoamicin biosynthetic gene cluster (BGC) XabABCD from Xenorhabdus doucetiae, resulting in the generation of 25 novel xenoamicin derivatives.