Contrastive Representation Learning for Single Cell Phenotyping in Whole Slide Imaging of Enrichment-free Liquid Biopsy
Contrastive Representation Learning for Single Cell Phenotyping in Whole Slide Imaging of Enrichment-free Liquid Biopsy
Naghdloo, A.; Tessone, D.; Mandya Nagaraju, R.; Zhang, B.; Kang, J.; Li, S.; Oberai, A.; Hicks, J. B.; Kuhn, P.
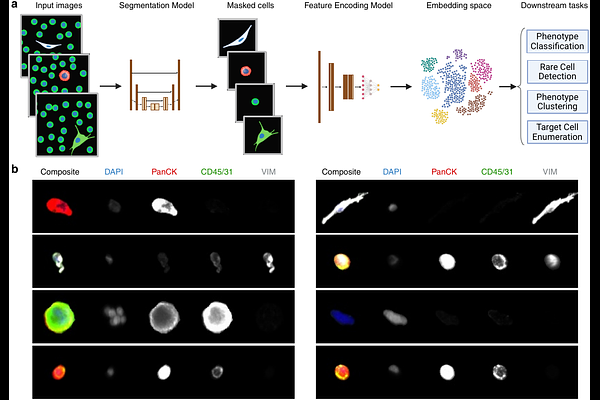
AbstractTumor-associated cells derived from a liquid biopsy are promising biomarkers for cancer detection, diagnosis, prognosis, and monitoring. However, their rarity, heterogeneity and plasticity make precise identification and biological characterization challenging for clinical utility. Enrichment-free approaches using whole slide imaging of all circulating cells offer a comprehensive and unbiased strategy for capturing the full spectrum of tumor-associated cell phenotypes. However, current analysis methods often depend on engineered features and manual expert review, making them sensitive to technical variations and subjective biases. These limitations highlight the need for a better feature representation to improve performance and reproducibility of applications in large-scale patient cohort analyses. In this study, we present a deep contrastive learning framework for learning features of all circulating cells, enabling robust identification and stratification of single cells in whole slide immunofluorescence microscopy images. We demonstrate performance of learned features in classification of diverse cell phenotypes in the liquid biopsy, achieving an accuracy of 92.64%. We further demonstrate that learned features improve performance in downstream applications such as outlier detection and clustering. Lastly, our feature representation enables automated identification and enumeration of distinct rare cell phenotypes, achieving average F1-score of 0.93 across cell lines mimicking circulating tumor cells and endothelial cells in contrived samples and average F1-score of 0.858 across CTC phenotypes in clinical samples. This workflow has significant implications for scalable analysis of tumor-associated cellular biomarkers in clinical prognosis and personalized treatment strategies.