MINN: A METABOLIC-INFORMED NEURAL NETWORK FOR INTEGRATING OMICS DATA INTO GENOME-SCALE METABOLIC MODELING
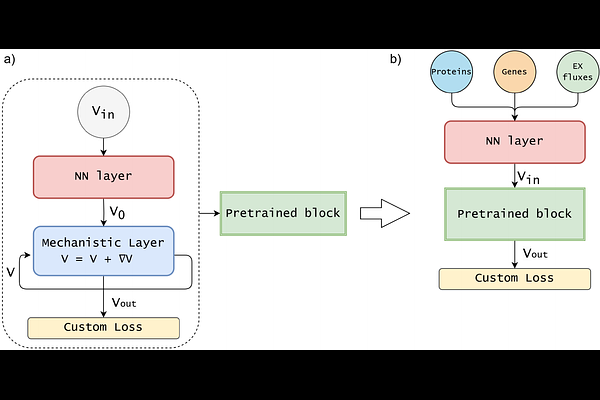
MINN: A METABOLIC-INFORMED NEURAL NETWORK FOR INTEGRATING OMICS DATA INTO GENOME-SCALE METABOLIC MODELING
Tazza, G.; Moro, F.; Ruggeri, D.; Teusink, B.; Vidacs, L.
AbstractThe understanding of cellular behavior relies on the integration of metabolism and its regulation. Multi-omics data provide a detailed snapshot of the molecular processes underpinning cellular functions and their regulation, describing the current state of the cell. While Machine Learning (ML) models can uncover complex patterns and relationships within these data, they require large datasets for training and often lack interpretability. On the other hand, mathematical models, such as Genome-Scale Metabolic Models (GEMs), offer a structured framework for analyzing the organization and dynamics of specific cellular mechanisms. At the same time, they do not allow for seamless integration of omics information. Recently, a new framework to embed GEMs in a neural network has been introduced: these hybrid models combine the strengths of mechanistic and data-driven approaches, offering a promising platform for integrating different data sources with mechanistic knowledge. In this study, we present a Metabolic-Informed Neural Network (MINN) that utilizes multi-omics data to predict metabolic fluxes in Escherichia coli, under different growth rates and gene knockouts. We test its performances against pure ML and parsimonious Flux Balance Analysis (pFBA), demonstrating its efficacy in improving prediction performances. We also highlight how conflicts can emerge between the data-driven and the mechanistic objectives, and we propose different solutions to mitigate them. Finally, we illustrate a strategy to couple the MINN with pFBA, enhancing the interpretability of the solution.