DiffDock-Glide: a hybrid physics-based and data-driven approach to molecular docking
DiffDock-Glide: a hybrid physics-based and data-driven approach to molecular docking
Herron, L.; Dakka, J.; Yao, K.; Shi, D.; Zhang, Y.; Jerome, S. V.
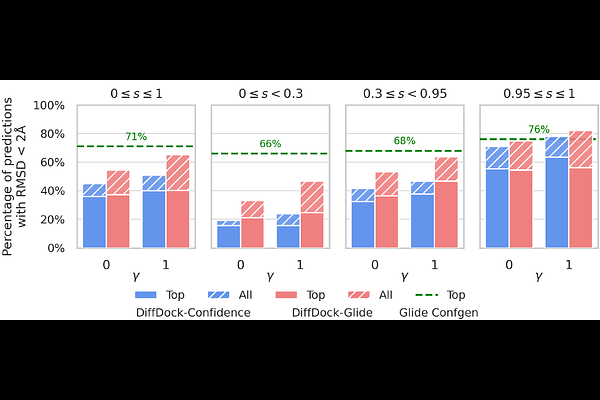
AbstractRecent years have seen a rise in applications of deep learning to problems in the molecular sciences. Among them, the diffusion model DiffDock stands out as a method for docking small molecules into protein binding sites. But DiffDock struggles to compete with conventional docking methods, especially for targets outside its training set. We develop a hybrid model called DiffDock-Glide which addresses some shortcomings of deep learning docking methods: it uses a modified generative process to generate samples within a binding pocket and the confidence model is replaced with Glide\'s post-docking minimization pipeline. We evaluate DiffDock-Glide on the Posebusters dataset and show improved sampling of near-native poses, especially for sequences without homologues in the training set. We also evaluate DiffDock-Glide\'s performance in virtual screening compounds from the DUD-E dataset against receptor structures generated by AlphaFold2 and report enrichment values that broadly surpass those from traditional Glide.