A novel bioinformatics approach reveals key transcription factors regulating type 1 diabetes-associated transcriptomes
A novel bioinformatics approach reveals key transcription factors regulating type 1 diabetes-associated transcriptomes
Sheikh, A.; Parvin, S.; Dipto, S.; Hossain, S.
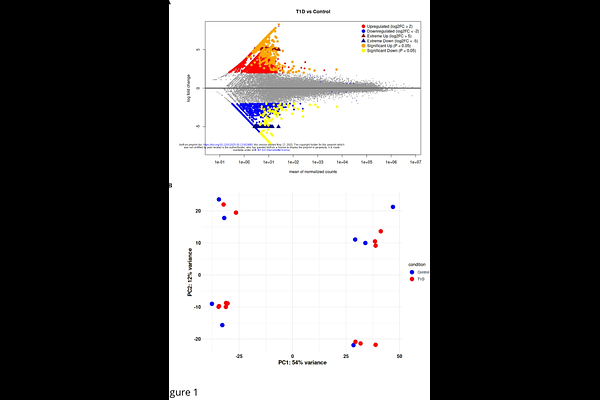
AbstractWe developed a novel bioinformatics pipeline that reveals key transcription factors (TFs) regulating type 1 diabetes transcriptomes by combining automated Python workflows, DESeq2-based differential expression analysis, motif enrichment, and genome-wide TF abundance profiling, a rare but powerful strategy that deepens our understanding of disease mechanisms. This approach not only uncovers TFs driving pathology but also quantifies TF abundance across multiple genomic loci, enabling rapid and precise monitoring of their occupancy. Analyzing 21 RNA sequencing datasets, including 13 from early-stage T1D patients and 8 matched controls, we identified nearly 6 000 differentially expressed genes; 1 900 met strict significance and fold-change criteria and 211 are previously uncharacterized transcripts that distinguish T1D from healthy samples. Pathway analysis highlighted disruptions in beta-cell signaling, ALK-linked drug responses, neurodegenerative processes, and cytoskeletal organization. Upstream motif analysis revealed enrichment of Myc/Max, AP-1, SP-1, TATA-box, and NF-{kappa}B binding sites in upregulated genes, confirming their central role in the T1D transcriptome. By placing TFs at the core of our discovery platform, this work uncovers novel molecular drivers of T1D and identifies actionable biomarkers and therapeutic targets for personalized treatment.