Macroscopic Analyses of RNA-Seq Data to Reveal Chromatin Modifications in Aging and Disease
Macroscopic Analyses of RNA-Seq Data to Reveal Chromatin Modifications in Aging and Disease
Mahajan, A.; Ratti, F.; Wang, B.; El-Samad, H.; Kaufman, J. H.; Gopalakrishnan, V.
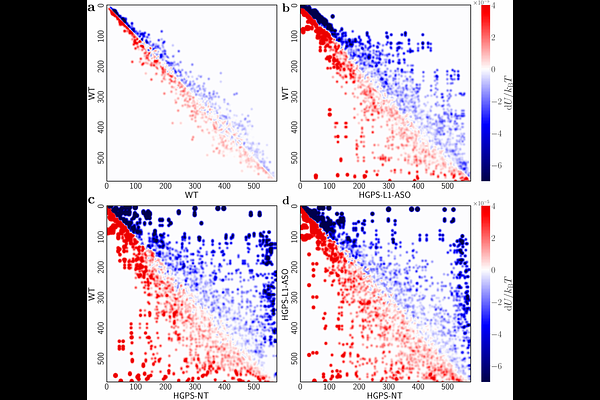
AbstractRegulation of gene expression is fundamental for proper cellular function, and is constrained by the local chromatin environment of each gene, which varies spatially along the chromosome and is shaped by epigenetic modifications. Epigenetic modifications induce changes in the local chromatin structure, which can influence gene expression, by affecting the accessibility of DNA to transcription factors. Such changes are particularly relevant in aging and genetic disorders like Hutchinson-Gilford Progeria Syndrome (HGPS) and Werner Syndrome (WRN), where altered chromatin structure contributes to disease pathology. In this study, we analyze RNA-seq data using macroscopic metrics designed to be explicitly sensitive to chromatin modifications. The first metric, intra-chromosomal gene correlation length, measures spatial correlations in gene expressions along the chromosome. The second metric employs an energy landscape model based on the Arrhenius equation to estimate the energetic barriers associated with chromatin state transitions. We apply these metrics to various aging-related datasets, demonstrating their sensitivity to changes in the chromatin structure and the interpretability of the resulting outputs. The intra-chromosomal gene correlation length is particularly effective in quantifying changes in RNA-seq profiles due to increased chromatin accessibility during aging (and conversely, reduced accessibility due to treatment). This metric not only accurately distinguishes cell states, but also provides insight into the direction of aging. For instance, our observations on the effects of anti-sense oligonucleotide (ASO) treatment align with the existing literature, demonstrating that ASO partially restores chromatin structure in diseased cells. They additionally quantify the more pronounced effects in HGPS compared to WRN. The barrier energy landscape further extends this capability by offering a framework for understanding the progressive degradation of the regulatory mechanisms. Together, these metrics provide robust screening tools that enhance our ability to exploit common measurements such as RNA-seq to derive new phenotypes such as chromatin dynamics on aging and disease, offering an alternative perspective that complements traditional analytical techniques and enriches our understanding of cellular states.