Erlotinib induces 3D genome rearrangements in lung cancer cells activating tumor suppressor genes through FOXA2-bound Epromoters
Erlotinib induces 3D genome rearrangements in lung cancer cells activating tumor suppressor genes through FOXA2-bound Epromoters
Swaminathan, G.; Cordero, J.; Guenther, S.; Graumann, J.; Braun, T.; Dobreva, G.; Barreto, G.
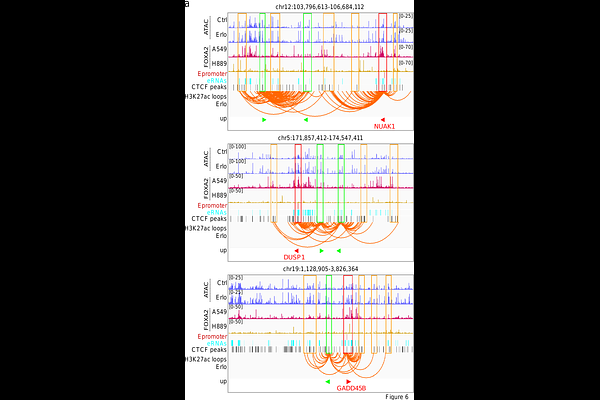
AbstractNon-small cell lung cancer (NSCLC) is the most frequent lung cancer (LC). While erlotinib is an epidermal growth factor (EGFR) tyrosine kinase inhibitor (TKI) used in the treatment of NSCLC, it remains unclear how this FDA-approved drug affects the genome. We performed integrative multi-omics studies in human pulmonary carcinoma cells to elucidate the epigenetic mechanisms induced by erlotinib. We identified 746 genes (including 34 tumor suppressor genes, TSG) that were upregulated after treatment with erlotinib or gefitinib (another EGFR-TKI). Interestingly, 45% of the upregulated genes (including 24 TSG) were in broad domains of the euchromatin histone mark H3K4me3, and 63% (including 26 TSG) exhibited reduced levels of the heterochromatin histone mark H3K27me3 after erlotinib treament. Further, H3K27ac-specific chromosome conformation capture-based methods revealed that erlotinib significantly increased number and length of chromatin loops between promoters of upregulated genes and active enhancers. We also detected augmented chromatin accessibility after erlotinib treatment at the promoters of upregulated genes, which correlated with binding of the transcription activator FOXA2. Remarkably, we identified gene clusters that seem to be upregulated by promoters with enhancer activity (Epromoters) enriched with FOXA2. The clinical relevance of our findings was confirmed by data from The Cancer Genome Atlas, showing significantly improved survival outcomes in LC patients with high levels of FOXA2 and/or the 34 TSG found upregulated by erlotinib. Our results establish 3D genome rearrangements as molecular mechanism mediating EGFR-TKI effects in NSCLC cells, supporting the design of more specific therapies for NSCLC targeting different chromatin features.