A whole genomic CRISPR-Cas9 screen identifies the amino acid transporter SLC43A1 (LAT3) as a major determinant of oxaliplatin sensitivity in colorectal cancer cells
A whole genomic CRISPR-Cas9 screen identifies the amino acid transporter SLC43A1 (LAT3) as a major determinant of oxaliplatin sensitivity in colorectal cancer cells
Pawar, N. R.; Wade, H. M.; Jackson, Z.; Poungpeth, N.; Mitchell, A. V.; Jewell, C. P.; Chan, D.; Robey, R. W.; Batista, P. J.; Jenkins, L. M.; Gottesman, M. M.
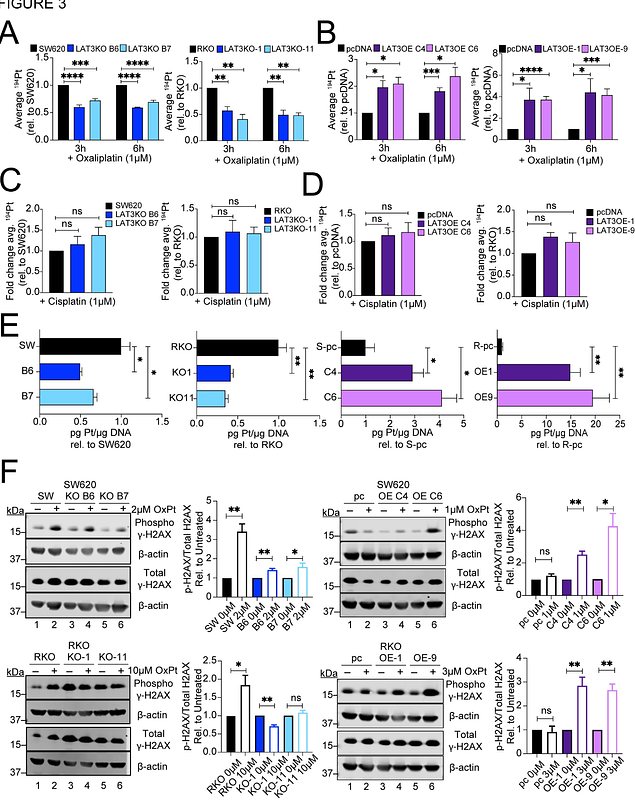
AbstractColorectal cancer (CRC) is the second leading cause of cancer deaths in the United States, with a five-year survival rate of 65%. Oxaliplatin was the first platinum drug shown to improve CRC patient outcomes and is now a common adjuvant therapy for advanced disease, yet 90% of patients develop resistance. Oxaliplatin was developed as a third-generation derivative of cisplatin, but recent evidence points to divergent modes of action. Here, genome-wide CRISPR activation and knockout screens were conducted to identify genetic changes that confer resistance to oxaliplatin in two CRC cell lines with distinct molecular backgrounds (SW620 and RKO). Guide RNAs corresponding to the neutral amino acid transporter SLC43A1 (LAT3) were the most significantly enriched in knockout screens and depleted in activation screens, suggesting a potential role for LAT3 in modulating oxaliplatin resistance. In vitro CRISPR knockout and overexpression of LAT3 in SW620 and RKO cell lines confirm increased resistance or sensitivity to oxaliplatin, respectively. Further analysis demonstrates that increased LAT3 levels corrrelate with increased intracellular levels of oxaliplatin, increased levels of DNA-platinum adducts and DNA damage, demonstrating that enhanced LAT3-mediated uptake of oxaliplatin can exert its expected mechanism of action and induce cytotoxicity. These findings may lead to a better understanding of oxaliplatin\'s mode of action in CRC and can provide new insights into the interplay between essential nutrient uptake and drug transport.