An ultrasensitive spatial tissue proteomics workflow exceeding 100 proteomes per day
An ultrasensitive spatial tissue proteomics workflow exceeding 100 proteomes per day
Klingeberg, M.; Krisp, C.; Fritzsche, S.; Schallenberg, S.; Hornburg, D.; Coscia, F.
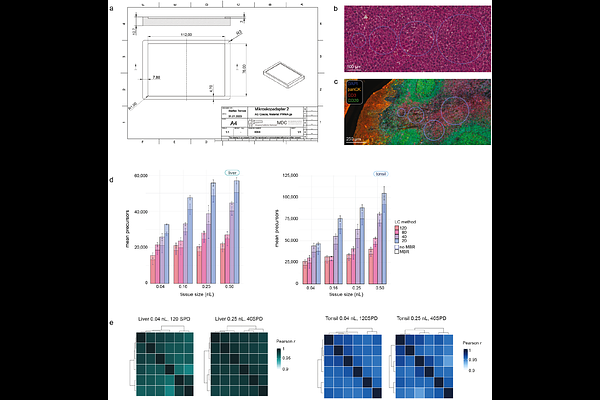
AbstractAchieving high-resolution spatial tissue proteomes requires careful balancing and integration of optimized sample processing, chromatography, and MS acquisition. Here, we present an advanced cellenONE protocol for loss-reduced tissue processing and compare all Evosep ONE Whisper Zoom gradients (20, 40, 80, and 120 samples per day), along with three common DIA acquisition schemes on a timsUltra AIP mass spectrometer. We found that tissue type was as important as gradient length and sample amount in determining proteome coverage. Moreover, the benefit of increased tissue sampling was gradient- and dynamic range-dependent. Analyzing mouse liver, a high dynamic range tissue, over tenfold more tissue led to only ~30% higher protein identification for short gradients (120 SPD and 80 SPD). However, even the lowest tested tissue amount (0.04 nL) yielded 3,200 reproducibly quantified proteins for the 120 SPD method. Longer gradients (40 SPD and 20 SPD) instead significantly benefited from more tissue sampling, quantifying over 7,500 proteins from 0.5 nL of tonsil T-cell niches. Finally, we applied our workflow to a rare squamous cell carcinoma of the oral cavity, uncovering disease-associated pathways and proteins with high intratumoral heterogeneity. Our study demonstrates that more than 100 high-quality spatial tissue proteomes can be prepared and acquired daily, laying a strong foundation for cohort-size spatial tissue proteomics in translational research.