HEAL-KGGen: A Hierarchical Multi-Agent LLM Framework with Knowledge Graph Enhancement for Genetic Biomarker-Based Medical Diagnosis
HEAL-KGGen: A Hierarchical Multi-Agent LLM Framework with Knowledge Graph Enhancement for Genetic Biomarker-Based Medical Diagnosis
Zuo, K.; Zhong, Z.; Huang, P.; Tang, S.; Chen, Y.; Jiang, Y.
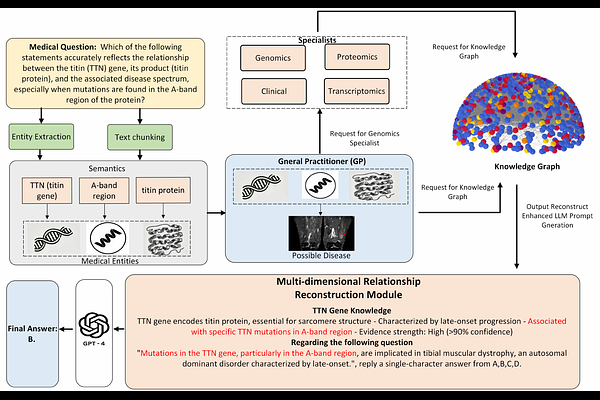
AbstractThe discovery and validation of genetic biomarkers across diverse diseases demand intelligent systems capable of integrating complex multi-omics data with clinical relevance. We introduce HEAL-KGGen, an end-to-end framework that enhances Large Language Models (LLMs) through a hierarchical multi-agent architecture and an automatically constructed medical knowledge graph. The system includes a General Practitioner (GP) agent for initial biomarker triage and specialist agents for genomics, transcriptomics, proteomics, and clinical interpretation. The core innovation of HEAL-KGGen lies in its dynamic knowledge graph pipeline, which combines entity extraction based on patterns and semantics, ontology-aligned normalization (using UMLS, MeSH, SNOMED CT), and the construction of multi-source relationships from biomedical databases and literature. Retrieved subgraphs are transformed into contextual prompts that guide LLM reasoning via structured, explainable pathways. Our experiments show that HEAL-KGGen significantly improves question-answering accuracy across multiple mainstream large language models, with the highest improvement observed on Claude 3.5 Sonnet, achieving a 43.75% increase in accuracy. These findings confirm the value of domain-specific graph knowledge in advancing LLM performance for genetic and molecular diagnostics.