A streamlined and comprehensive protocol for the generation and multi-omic analysis of human monocyte-derived macrophages
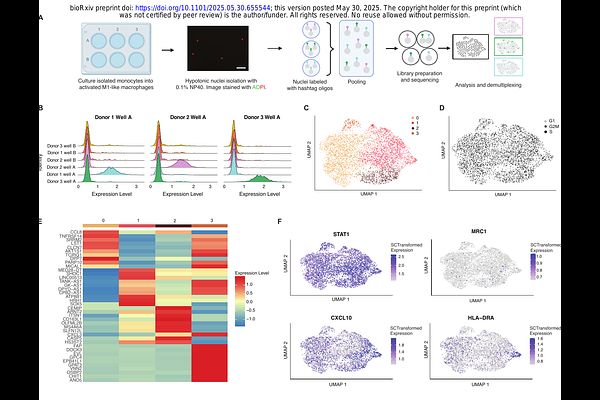
A streamlined and comprehensive protocol for the generation and multi-omic analysis of human monocyte-derived macrophages
Palmer, O. G.; Perreard, L.; Kolling, F. W.; Pioli, P. A.; Goods, B. A.
AbstractMacrophages serve both as a first line of defense against invading pathogens and mediate tissue homeostasis. These cells are inherently responsive and heterogeneous and lie on a spectrum of activation states book-ended by M1-like (inflammatory) and M2-like (anti-inflammatory) extremes. The study of human macrophages is necessary to unravel the complex signals and environmental cues that these cells integrate to create their varied phenotypes. In vitro protocols to differentiate human monocytes into macrophages use many distinct activation stimuli at variable concentrations and for differing durations of treatment that can impact macrophage fate. These variations can make it challenging to reproduce findings and compare datasets across research environments. Additionally, few protocols to date have performed rigorous characterization with input material from frozen peripheral blood mononuclear cells (PBMCs). This is important since the availability of fresh blood can often be limiting and can lead to a loss of standardized procedures, particularly for cell therapy applications. Here, we have developed a comprehensive protocol to generate human macrophages from monocytes where we rigorously characterize the impact of differentiation conditions and polarization conditions on human macrophages. We provide a detailed protocol for their characterization using several omics readouts, including their cytokine production and transcriptomes. We also perform depolarization experiments to determine durability of macrophage immunophenotype post-removal of polarizing stimuli. Finally, we demonstrate that nuclei can be isolated and profiled by snRNA-seq directly from macrophages in culture, alleviating the need to detach these adherent cells for downstream multi-ome analyses. Taken together, we provide a comprehensive, detailed and streamlined procedure for the differentiation and characterization of human macrophages from monocytes isolated from frozen PBMCs. This is important for enabling the study of macrophages in a more systematic way from biobanked material.