CMiNet: An R Package and User-Friendly Shiny App for Constructing Consensus Microbiome Networks
CMiNet: An R Package and User-Friendly Shiny App for Constructing Consensus Microbiome Networks
Aghdam, R.; Solis-Lemus, C.
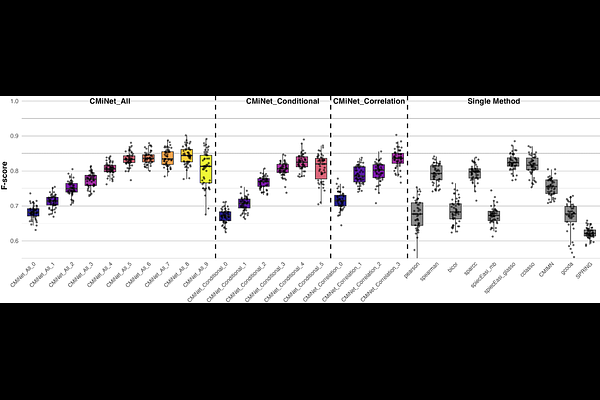
AbstractMicrobial networks offer critical insights into community structure, ecological interactions, and host microbe dynamics. However, constructing reliable microbiome networks remains challenging due to variability among existing inference methods, limited overlap between networks, and the absence of a gold standard for validation. We developed CMiNet https://cminet.wid.wisc.edu), an interactive Shiny application and R package that enables consensus microbiome network construction by integrating up to ten widely used inference algorithms. CMiNet supports both correlation-based and conditional dependence-based methods and provides users with flexible options to construct individual or consensus networks across different approaches. CMiNet employs a consensus-based filtering strategy that retains only edges supported by multiple methods, leading to more robust network structures that better reflect underlying biological interactions. This approach enhances reproducibility, minimizes method-specific biases, and improves biological interpretability. Its user-friendly interface allows researchers to upload data, customize network construction parameters, visualize networks interactively, and export results without requiring programming expertise. We demonstrated CMiNet using both gut and soil microbiome datasets. In the soil microbiome application, robust network construction enabled the consistent identification of disease-associated taxa. Specifically, we identified a set of key taxa, including Ktedonobacteria, Acidobacteriae, Vicinamibacteria, MB-A2-108, Planctomycetes, and Anaerolineae, selected by multiple independent methods.