ProtFun: A Protein Function Prediction Model Using Graph Attention Networks with a Protein Large Language Model
ProtFun: A Protein Function Prediction Model Using Graph Attention Networks with a Protein Large Language Model
Talo, M.; Bozdag, S.
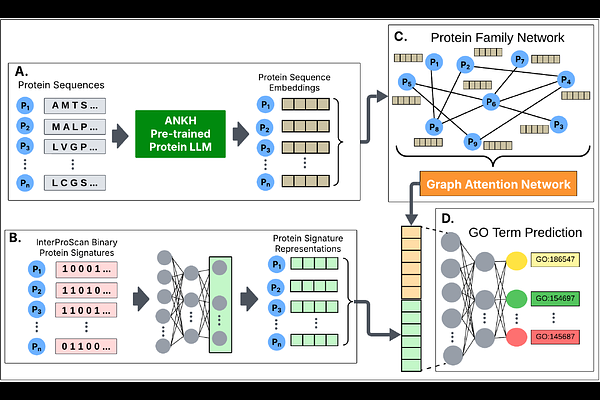
AbstractUnderstanding protein functions facilitates the identification of the underlying causes of many diseases and guides the research for discovering new therapeutic targets and medications. With the advancement of high throughput technologies, obtaining novel protein sequences has been a routine process. However, determining protein functions experimentally is cost- and labor-prohibitive. Therefore, it is crucial to develop computational methods for automatic protein function prediction. In this study, we propose a multi-modal deep learning architecture called ProtFun to predict protein functions. ProtFun integrates protein large language model (LLM) embeddings as node features in a protein family network. Employing graph attention networks (GAT) on this protein family network, ProtFun learns protein embeddings, which are integrated with protein signature representations from InterPro to train a protein function prediction model. We evaluated our architecture using \\textcolor{red}{three} benchmark datasets. Our results showed that our proposed approach outperformed current state-of-the-art methods for most cases. An ablation study also highlighted the importance of different components of ProtFun. The data and source code of ProtFun is available at https://github.com/bozdaglab/ProtFun under Creative Commons Attribution Non Commercial 4.0 International Public License.