Unveiling Genomic Rearrangements in Engineered iPSC Lines by Optical Genome Mapping
Unveiling Genomic Rearrangements in Engineered iPSC Lines by Optical Genome Mapping
Finlay, D.; Hor, P.; Goldenson, B. H.; Li, X.-H.; Murad, R.; Kaufman, D. S.; Vuori, K.
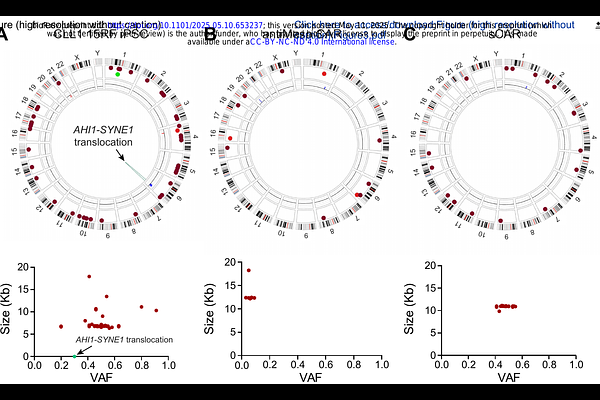
AbstractWe demonstrate here the use of optical genome mapping (OGM) to detect genetic alterations arising from gene editing by various technologies in human induced pluripotent stem cells (iPSCs). OGM enables an unbiased and comprehensive analysis of the entire genome, allowing the detection of genomic structural variants (SVs) of all classes with a quantitative variant allele frequency (VAF) sensitivity of 5%. In this pilot study, we conducted a comparative dual analysis between the parental iPSCs and the derived cells that had undergone gene editing using various techniques, including transposons, lentiviral transduction, and CRISPR-Cas9-mediated safe harbor locus insertion at the adeno-associated virus integration site 1 (AAVS1). These analyses demonstrated that iPSCs that had been edited using transposons or lentiviral transduction resulted in a high number of transgene insertions in the genome. In contrast, CRISPR-Cas9 technology resulted in a more precise and limited transgene insertion, with only a single target sequence observed at the intended locus. These studies demonstrate the value of OGM to detect genetic alterations in engineered cell products and suggests that OGM, together with DNA sequencing, could be a valuable tool when evaluating genetically modified iPSCs for research and therapeutic purposes.