Explainable Machine Learning Identifies Dosage Compensation Factors in Aneuploid Human Cancer Cells
Explainable Machine Learning Identifies Dosage Compensation Factors in Aneuploid Human Cancer Cells
Heller, E. M.; Barthel, K.; Raschle, M.; Schukken, K.; Sheltzer, J.; Storchova, Z.
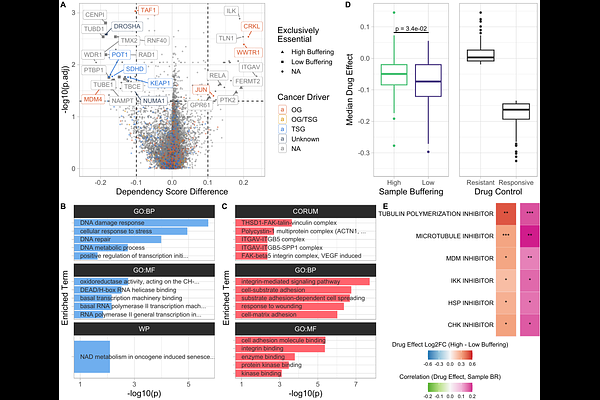
AbstractAneuploidy, a hallmark of cancer, alters chromosome copy numbers and with that the abundance of hundreds of proteins. Evidence suggests that levels of proteins encoded on affected chromosomes are often buffered toward their abundances observed in diploids. Despite its prevalence, the molecular mechanisms driving this protein dosage compensation remain largely unknown. It is unclear whether all proteins are buffered similarly, what factors determine buffering, and whether dosage compensation varies across different cell lines or tumor types. Moreover, its potential adaptive advantage and therapeutic relevance remain unexplored. We established a novel approach to quantify protein dosage buffering in a gene copy number-dependent manner, showing that dosage compensation is widespread but variable in cancer samples. By developing multifactorial machine learning models, we identify gene dependency, protein complex participation, haploinsufficiency, and mRNA decay as key predictors of buffering. We show that dosage compensation affects oncogenic potential, and that higher buffering correlates with reduced proteotoxic stress and increased drug resistance. These findings highlight protein dosage compensation as a crucial regulatory mechanism with therapeutic potential in aneuploid cancers.