Efficient genetic code expansion tools enable in vivo study of lysine acetylation in non-model bacteria
Efficient genetic code expansion tools enable in vivo study of lysine acetylation in non-model bacteria
Van Fossen, E. M.; Stephenson, M.; Frank, A.; Akella, S.; Wooldridge, R.; Wilson, A.; You, Y.; Nakayasu, E.; Egbert, R.; Elmore, J. R.
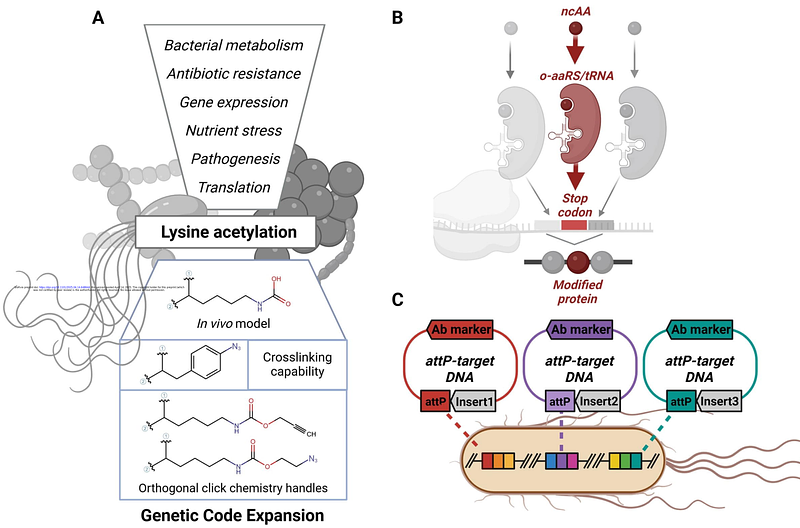
AbstractRecent proteomic advancements have revealed widespread N{varepsilon}-lysine acetylation in pathways governing pathogenicity, metabolism, and antibiotic resistance in bacteria. The spontaneous, non-specific nature of this modification in prokaryotes obscures its biological role, necessitating prokaryotic specific in vivo interrogation systems. Genetic Code Expansion (GCE) offers a powerful method to investigate the roles and regulation dynamics of acetyl-lysine in vivo with the precise incorporation of a suite of non-canonical amino acids, including acetyl-lysine analogs. However, its use has been largely restricted to Escherichia coli strains due to challenges associated with implementation and optimization of the technology in more diverse bacterial strains. Here, we present a bacterial host-agnostic, readily optimizable genetic code expansion platform designed to site-specifically incorporate non-canonical amino acids (ncAAs) into target proteins within living bacteria. We further demonstrate the versatility of this technology by showcasing, for the first time, the successful incorporation of acetyl-lysine in a non- E. coli bacterium.