Coreset-based logistic regression for atlas-scale cell type annotation
Coreset-based logistic regression for atlas-scale cell type annotation
Li, D.; Ko, Y. K.; Canzar, S.
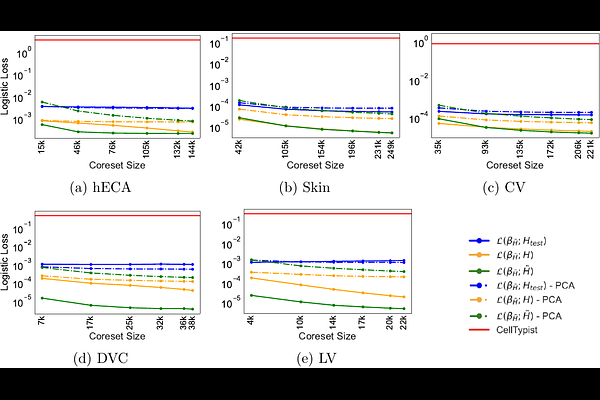
AbstractLogistic regression provides a powerful tool for the predictive analysis of single cell and spatial omics data. It has been shown to outperform more complex models on fundamental tasks such as the selection of marker genes and the annotation of cell types. Training logistic regression models on increasingly large, publicly available single-cell datasets opens up opportunities to build accurate and robust cell type prediction models but poses major computational challenges. Here, we adapt and extend the theory of coresets for logistic regression. In particular, we compute a previously introduced classification complexity measure using linear programming to identify single-cell datasets which admit coresets, i.e. random samples of cells, that preserve logistic loss. We show that applying this complexity measure requires a prior dimensionality reduction by, e.g., principle component analysis, and prove that logistic loss is preserved under this linear transformation. In experiments on several cell atlases, we compared our method to CellTypist, a logistic regression-based immune cell atlas and scMulan, a pre-trained language model, to demonstrate that theoretical guarantees translate to an accurate cell type label transfer method that scales to atlas-scale datasets.