Gene-embedding-based prediction and functional evaluation of perturbation expression responses with PRESAGE
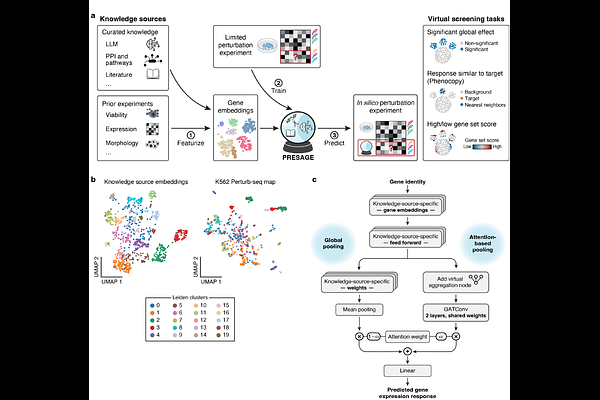
Gene-embedding-based prediction and functional evaluation of perturbation expression responses with PRESAGE
Littman, R.; Levine, J.; Maleki, S.; Lee, Y.; Ermakov, V.; Qiu, L.; Wu, A.; Huang, K.; Lopez, R.; Scalia, G.; Biancalani, T.; Richmond, D.; Regev, A.; Hütter, J.-C.
AbstractUnderstanding the impact of genetic perturbations on cellular behavior is crucial for biological research, but comprehensive experimental mapping remains infeasible. We introduce PRESAGE (Perturbation Response EStimation with Aggregated Gene Embeddings), a simple, modular, and interpretable framework that predicts perturbation-induced expression changes by integrating diverse knowledge sources via gene embeddings. PRESAGE transforms gene embeddings through an attention-based model to predict perturbation expression outcomes. To assess model performance, we introduce a comprehensive evaluation suite with novel functional metrics that move beyond traditional regression tasks, including measures of accuracy in effect size prediction, in identifying perturbations with similar expression profiles (phenocopy), and in prediction of perturbations with the strongest impact on specific gene set scores. PRESAGE outperforms existing methods in both classical regression metrics and our novel functional evaluations. Through ablation studies, we demonstrate that knowledge source selection is more critical for predictive performance than architectural complexity, with cross-system Perturb-seq data providing particularly strong predictive power. We also find that performance saturates quickly with training set size, suggesting that experimental design strategies might benefit from collecting sparse perturbation data across multiple biological systems rather than exhaustive profiling of individual systems. Overall, PRESAGE establishes a robust framework for advancing perturbation response prediction and facilitating the design of targeted biological experiments, significantly improving our ability to predict cellular responses across diverse biological systems.