Learning Genetic Perturbation Effects with Variational Causal Inference
Learning Genetic Perturbation Effects with Variational Causal Inference
Liu, E.; Zhang, J.; Uhler, C.
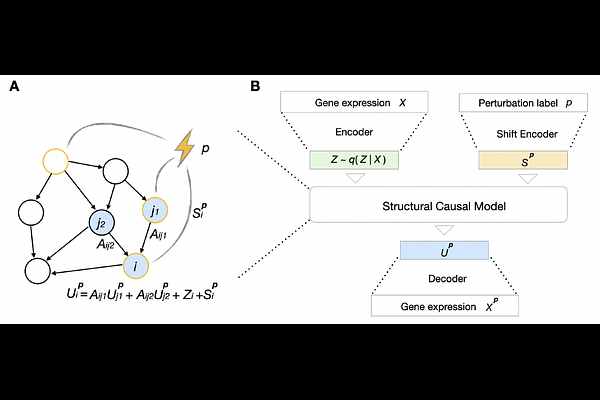
AbstractAdvances in sequencing technologies have enhanced the understanding of gene regulation in cells. In particular, Perturb-seq has enabled high-resolution profiling of the transcriptomic response to genetic perturbations at the single-cell level. This understanding has implications in functional genomics and potentially for identifying therapeutic targets. Various computational models have been developed to predict perturbational effects. While deep learning models excel at interpolating observed perturbational data, they tend to overfit and may not generalize well to unseen perturbations. In contrast, mechanistic models, such as linear causal models based on gene regulatory networks, hold greater potential for extrapolation, as they encapsulate regulatory information that can predict responses to unseen perturbations. However, their application has been limited to small studies due to overly simplistic assumptions, making them less effective in handling noisy, large-scale single-cell data. We propose a hybrid approach that combines a mechanistic causal model with variational deep learning, termed Single Cell Causal Variational Autoencoder (SCCVAE). The mechanistic model employs a learned regulatory network to represent perturbational changes as shift interventions that propagate through the learned network. SCCVAE integrates this mechanistic causal model into a variational autoencoder, generating rich, comprehensive transcriptomic responses. Our results indicate that SCCVAE exhibits superior performance over current state-of-the-art baselines for extrapolating to predict unseen perturbational responses. Additionally, for the observed perturbations, the latent space learned by SCCVAE allows for the identification of functional perturbation modules and simulation of single-gene knockdown experiments of varying penetrance, presenting a robust tool for interpreting and interpolating perturbational responses at the single-cell level.